Adaptive Introgression 2024
Identification of a Novel Leaf Rust Resistance Gene
Leaf rust, caused by Puccinia triticina (Pt), is one of the most common wheat diseases globally. Pt is prevalent in areas with warm to high temperatures and high moisture, and Pt is highly diverse in pathogenicity with 40 to 60 races identified in the United States alone each year. The ability of Pt to rapidly evolve new pathotypes and large-scale production of urediniospores that can disperse over long distances create challenges for disease management. Leaf rust typically reduces kernel weight and number per spike, causing 5%-15% yield losses depending on the crop developmental stage at initial infection. Host resistance is the most efficient, cost-effective and environmentally friendly approach to managing leaf rust. Unfortunately, most known leaf rust resistance genes have lost effectiveness in the Great Plains. Therefore, novel resistance genes that can be rapidly introgressed (transferred) into locally adapted cultivars are urgently needed.
The Oklahoma State University Wheat Improvement Team identified the new leaf rust resistance gene Lr622111. Lr62211 that was discovered in the Iranian wheat landrace PI 622111 (Figure 1), which confers a wide spectrum of resistance to current representative Pt races in the U.S. (Table 1) and was delimited to a 0.4 Mb interval on the short arm of chromosome 5B (Figure 2). Further, an allelism test indicated that Lr 622111 is allelic to Lr52. Since PI 622111 reacted differently from the Lr52 donor to Pt races collected from Oklahoma, Lr622111 is considered a new Lr52 allele.

Figure 1. Responses of PI 622111 (left) and Yuanyu 3 (right) to Pt race Pt52-2.
| Infection type | PI 622111 | Thatcher | TAM 110 |
|---|---|---|---|
| BBBDB* | ;1- | 3+4 | |
| KFBJG | ; | 3+4 | |
| MPPSD | ;1- | 3_4 | |
| TCRKG | ;1- | 3+4 | |
| TNBJS | ;1- | 3+4 | |
| TBBGS | ;1- | 3+ | |
| TFTSB | ; | 3+ | |
| 0 | |||
| MBDSD | ; | 3+4 | |
| MJBJG | ;1- | 3+4 | |
| MCTNB | ; | 3+ | |
| MHDSB | ; | 3+ | |
| MNPSD | ; | 3+ | |
| SBDGG | ; | ;23 (X) | |
| TFBGQ | ; | 3+ | |
| TNRJJ | ; | 3+ | |
| PBLRG | ; | ;23 (X) |
* Infection type (IT) was scored 10 days after inoculation: Plants with IT “0” (immune response); “;” (hypersensitive fleck), “1” (small necrotic uredinia), and “2” (small size uredinia with prominent chlorosis) were considered resistant, and plants with IT “3” (moderate size uredinia lacking necrosis or chlorosis) to “4” (large uredinia) were considered susceptible. Plants that had mixed IT were described with the most common IT first. Symbols “–” and “ + ” denoted smaller or larger uredinia, respectively.
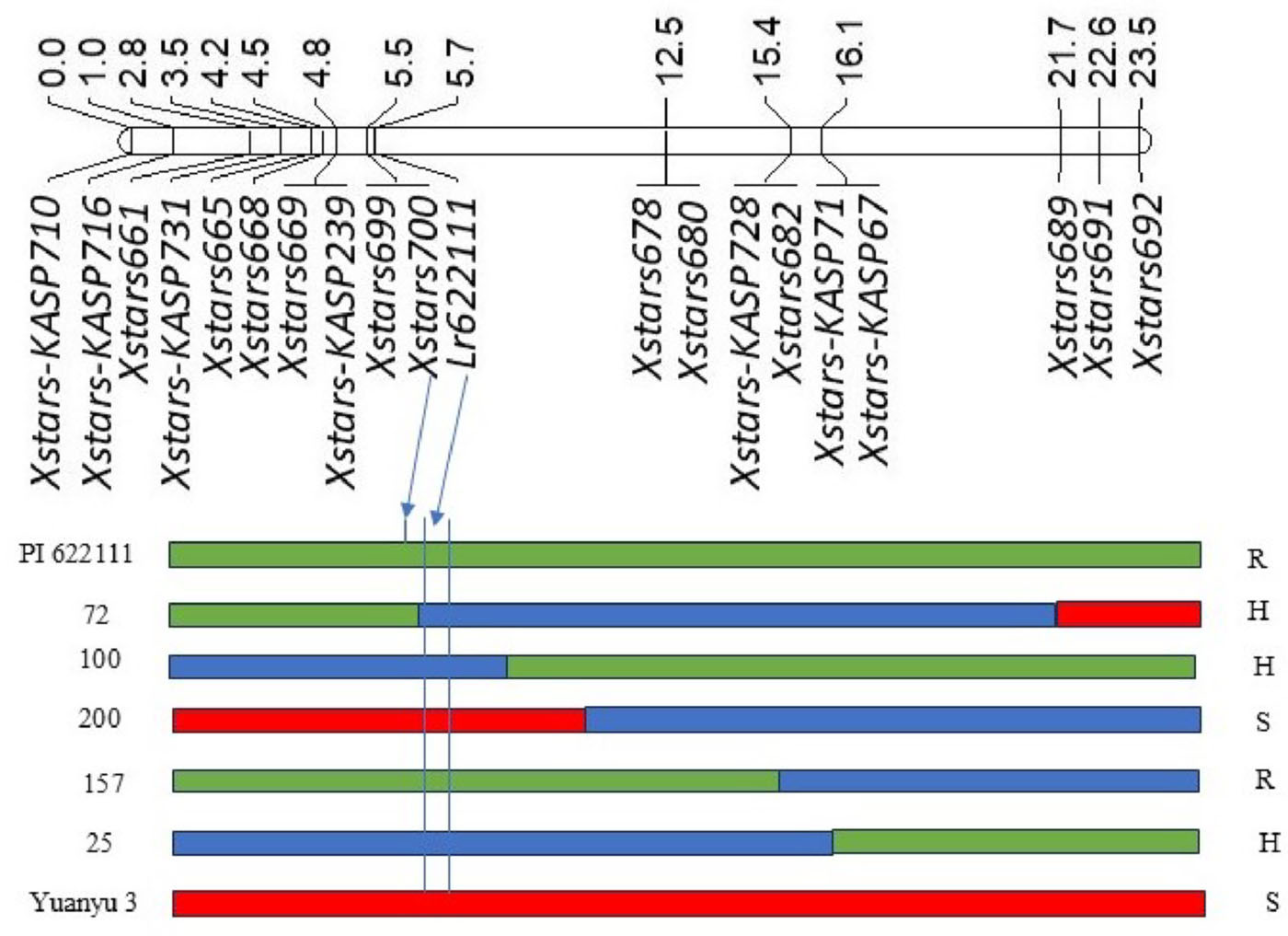
Figure 2. Linkage map showing genomic location of Lr622111 in the mapping population from PI 622111 × Yuanyu 3, and graphic genotype and phenotype of PI 622111, Yuanyu 3 and five F2:3 lines (72, 100, 200, 157 and 25). Molecular markers and genetic distances are below and above the linkage map, respectively. Green, red and blue bars represent PI 622111, Yuanyu 3 and heterozygous genotypes. R, S and H indicate resistant, susceptible and heterozygous phenotypes. The lines carrying resistant (green), susceptible (red) and heterozygous (blue) alleles at Lr622111 (between the two vertical bars) showed resistant, susceptible and heterozygous phenotypes.
The resistant allele at Xstars-KASP239, a kompetitive allele specific PCR (KASP) marker that is 0.9 cM distal to Lr622111, is uniquely found in Iranian landraces. The Lr52 donor GSTR 422 and Iranian landraces PI 621674, PI 622129, PI 622246 and PI 622066 carry the resistant allele, while all 86 U.S. cultivars and breeding lines carry the alternative allele (Figure 3). Therefore, Xstars-KASP239 can be widely used to tag Lr622111 in U.S. wheat breeding programs because polymorphism is expected between PI 622111 and U.S. cultivars.
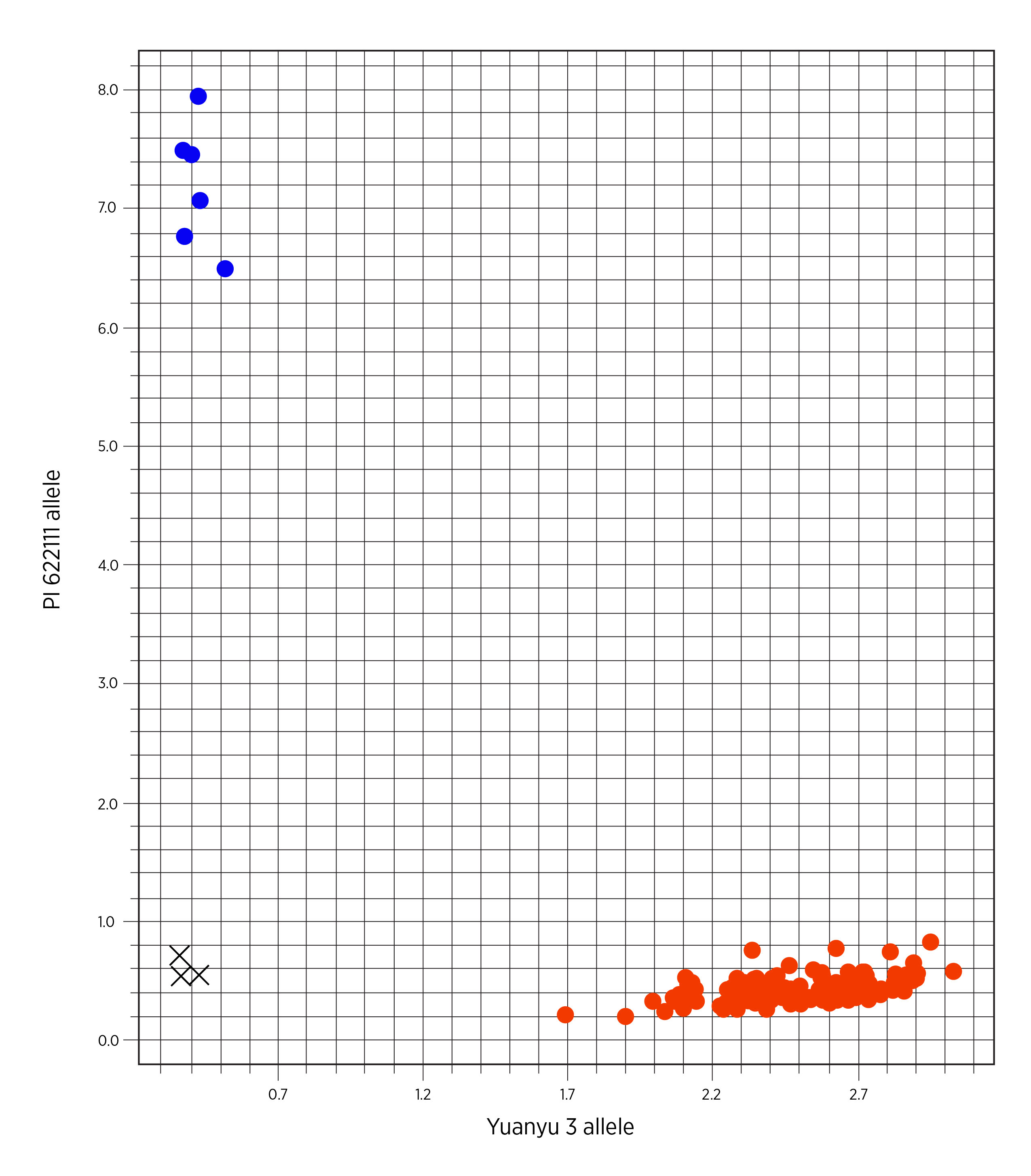
Figure 3. Allele distributions of PI 622111, Yuanyu 3, GSTR 422, PI 621674, PI 622129, PI 622246, PI 622066 and a set of 86 U.S. hard winter wheat cultivars/breeding lines from the 2019 Hard Winter Regional Performance Nursery at KASP marker Xstars-KASP239. Blue spot, red spot, and black check mark “X” represent PI 622111 allele, Yuanyu 3 allele and negative control (water). GSTR 422, the donor of Lr52, and Iranian landraces PI 622111, PI 621674, PI 622129, PI 622246 and PI 622066 carry the resistant allele (blue), whereas Yuanyu 3 and all 86 U.S. cultivars carry the susceptible allele at Xstars-KASP239 (red).
Discovery and Characterization of the Greenbug Resistance Gene Gb49921
Greenbug (Schizaphis graminum; aka Rondani) is a major insect pest that poses a serious threat to wheat production worldwide. Since the first report of greenbug infestation in Virginia in 1884, greenbug outbreaks have recurred every five to 10 years in the Great Plains, leading to significant economic losses. Greenbug infestation, especially at early growth stages, can reduce wheat yields by 40%-50% if left uncontrolled. Host resistance is preferred for managing greenbug, and nine genes (Gb1-Gb9) have been permanently designated. However, the continuous emergence of new greenbug biotypes in the field makes it urgent to identify novel greenbug resistance genes in wheat.
The WIT identified a new greenbug resistance gene, designated Gb49921, in the spring wheat breeding line BW 49921. Genetic analysis delimited Gb9 to a 0.5-Mb interval flanked by KASP markers Stars-KASP419 (598.8 Mb) and Stars-KASP1061 (599.3 Mb) on 7DL (Figure 4). Gb49921 was 1.0 cM distal to Stars-KASP419 and 0.6 cM proximal to Stars-KASP1061. Allelism tests indicated that Gb49921 is not allelic to either Gb3 or Gb8. Gb49921 confers high resistance to greenbug biotypes C, E, H, I and K and moderate resistance to TX1 (Table 2). The KASP markers developed in this study can be used for marker-assisted selection of Gb49921 in wheat breeding programs.

Figure 4. Genetic (left) and physical (right) maps showing genomic location of Gb49921. Marker names are shown at the right of the linkage map, and genetic distances in cM or physical positions in Mb of molecular markers are given on the left sides of genetic and physical maps. The resistant alleles of KASP markers flanking Gb49921 (such as Stars-KASP419 and Stars-KASP1061) are only found in the CWI49921, and these markers can be widely used in U.S. wheat breeding programs.
| wheat line | B | C | E | H | I | K | FL | TX1 |
|---|---|---|---|---|---|---|---|---|
| BW 49921 | S* | R | R | R | R | R | S | MR |
| CWI 76364 (Gb9) | S | R | R | R | R | R | S | R |
| Largo (Gb3) | S | R | R | R | R | R | S | S |
| CI 17959 (Gb4) | S | R | R | S | R | R | S | S |
| W7984 (Gb7) | S | S | R | S | R | R | S | S |
| PI 697274 (Gb8) | R | R | R | R | R | R | R | S |
| Jagalene | S | S | S | S | S | S | S | S |
*S, is susceptible, R is Resistant and MR is Moderately Resistant
Identification of a Gene Conferring Resistance to U.S. Russian Wheat Aphid Biotypes in the Iranian Landrace PI 625139
Russian wheat aphid (RWA, Diuraphis noxia Kurdjumov) is a highly invasive and destructive wheat pest evolving rapidly to overcome host resistance. RWA population size can rapidly increase via facultative parthenogenesis with apterous females giving birth to live nymphs, leading to up to 50.2%–82.9% of yield losses and 55.2%–76.5% of biomass reduction if left unchecked in RWA epidemics. Although several RWA resistance genes have been identified, most of them lost effectiveness because of RWA biotype shifts in the field. A recent field survey found RWA across the Great Plains, indicating potential RWA epidemics in this region in the future. Novel genes conferring resistance to multiple RWA biotypes are needed to sustain wheat production.
PI 625139 is a RWA-resistant landrace. The WIT discovered a gene conditioning resistance to all five U.S. RWA biotypes (RWA1, RWA2, RWA3/7, RWA6 and RWA8) in the proximal region of chromosome arm 7DS, and they mapped the gene (Dn625139) to a 2.6 cM interval between KASP marker Stars-KASP1007 (169.57 Mb) and SSR marker Stars1039 (195.27 Mb; Figure 5). Dn625139 co-segregated with the SSR marker Stars1029 (180.82 Mb). Dn625139 is valuable, and the molecular markers developed in this study warrant its rapid introgression into locally adapted cultivars to enhance wheat RWA resistance.
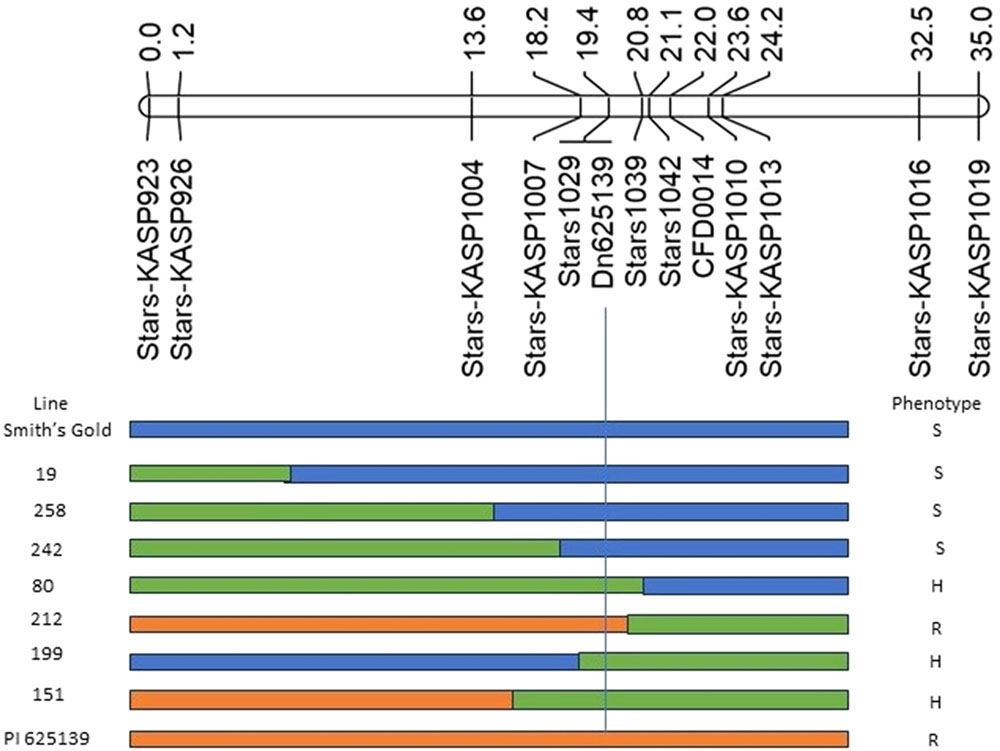
Figure 5. Partial 7DS linkage map showing the genomic location of Dn625139 and graphic presentation of Smith’s Gold, PI 625139 and seven selected F2:3 lines (19, 258, 242, 80, 212, 199 and 151) based on their genotypes and phenotypes. KASP markers and genetic distance (cM) were given below and above the linkage map. Orange, blue and green bars represent PI 625139, Smith’s Gold and heterozygous genotypes based on their F2 genotypic data. R, S and H represent homozygous resistant, homozygous susceptible and heterozygous phenotypes. The lines carrying resistant (yellow), susceptible (blue) and heterozygous (green) alleles at Dn625139 (vertical bars) showed resistant, susceptible and heterozygous phenotypes.
Pyramiding Novel Resistance Genes
WIT continued to transfer novel disease and pest-resistance genes into locally adapted lines. These included leaf rust resistance genes Lr81, Lr622111, Qlr.stars-1RS, QLr-Stars-2DS and QLr-Stars-6BL; powdery mildew resistance genes Pm59, Pm65 and Pm351817; and greenbug resistance genes Gb8and Gb9. A backcross population with Pm63 is currently being evaluated in fields. In addition, we initiated a new project aimed at transferring a Hessian fly resistance gene into Oklahoma wheat cultivars.
Acknowledgments
Mention of trade names or commercial products in this publication is solely to provide specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
