Adaptive Introgression 2022
Discovery and characterization of the novel leaf rust resistance gene Lr81
Leaf rust, caused by Puccinia triticina (Pt), is the most common and widespread disease affecting wheat production worldwide. Novel resistance sources are urgently needed in the Southern Great Plains of the United States because most leaf rust resistance genes originating from bread wheat are no longer effective in this region, and only a few known genes from wheat wild species still provide effective resistance. The transfer of these effective resistance genes into locally adapted cultivars typically takes 10 or more years and is often hindered by linkage drag, which is the co-introduction of unfavorable genetic traits linked with the resistance genes. This shortage of leaf rust resistance sources in this region is partially due to abundant variation in the pathogen population from which virulent races are selected rapidly after deployment of resistance genes in wheat cultivars.
The Oklahoma State University Wheat Improvement Team (WIT) identified a new leaf rust resistance gene, officially designated as Lr81, in the breeding line PI 470121 developed at the University of Zagreb in Croatia. This gene was delimited to a genomic region of 96,148 bp flanked by newly developed Kompetitive Allele specific PCR (KASP) markers Xstars-KASP320 (67,030,206 bp) and Xstars-KASP323 (67,132,354 bp) on the short arm of chromosome 2A in the Chinese Spring genome sequence (Fig. 1). Marker-assisted selection was subsequentially performed, leading to the selection of wheat lines stacked with four leaf rust resistance genes. Lr81 confers resistance to dominant Pt races and can be widely used to enhance leaf rust resistance in the U.S., particularly in Oklahoma.
In addition, a set of over 20 recombinant inbred line (RIL) or F2:3 populations has been evaluated for responses to commonly occurring Pt race Pt 52–2 (virulence phenotype designation MMPSD), and five of them are being genotyped to uncover the underlying leaf rust resistance genes.
*Note: PCR stands for polymerase chain reaction, referring to a laboratory technique for rapidly producing millions to billions of copies of a specific segment of DNA, which can then be studied in greater detail.
A novel gene conferring a wide spectrum resistance to powdery mildew
Powdery mildew, caused by the biotrophic fungus Blumeria graminis f. sp. tritici (Bgt), is a globally serious wheat disease causing up to 40% yield loss in susceptible wheat cultivars. There is higher occurrence and severity where intensive cultivation practices associated with high rainfall or supplemental irrigation and high levels of nitrogen fertilization have been adopted to maximize grain yield. With global warming, powdery mildew is predicted to be a more serious issue for wheat production, and powdery mildew resistance genes that can be easily transferred to locally adapted breeding lines are urgently needed.
WIT identified a new powdery mildew resistance gene in wheat accession PI 351817 that provides resistance to all 19 Bgt isolates collected from different wheat growing regions of the U.S. (Table 1). WIT evaluated responses of an F2 population and 237 F2:3 lines derived from the cross OK1059060-2C14 × PI 351817 to Bgt isolate OKS(14)-B-3-1. Genetic analysis indicated a single dominant gene, designated Pm351817, conferred powdery mildew resistance in PI 351817. The F2 population was further genotyped using a set of KASP markers, and Pm351817 was delimited to a 634-kb interval from 771,207,512 bp (Stars-KASP656) to 771,841,609 bp (Stars-KASP662) on chromosome arm 2AL in the Chinese Spring genome sequence IWGSC RefSeq v2.0 (Fig. 2). Allelism tests indicated that Pm351817 is a new Pm65 allele.
Pm351817 can be widely used to enhance powdery mildew resistance in the U.S., and KASP markers flanking Pm351817 have the potential to track the gene in wheat breeding populations. WIT immediately initiated an introgression program to distribute Pm351817 into contemporary germplasm adapted to Oklahoma and the southern U.S. Plains.
Also, one RIL and two additional F2:3 populations have been evaluated for responses to Bgt isolate OKS(14)-B-3-1, and genetic analyses are underway to characterize the powdery mildew resistance genes in these populations.
*Note: An allele is one of two or more alternative forms of a gene that arise by mutation and are found at the same place on a chromosome.
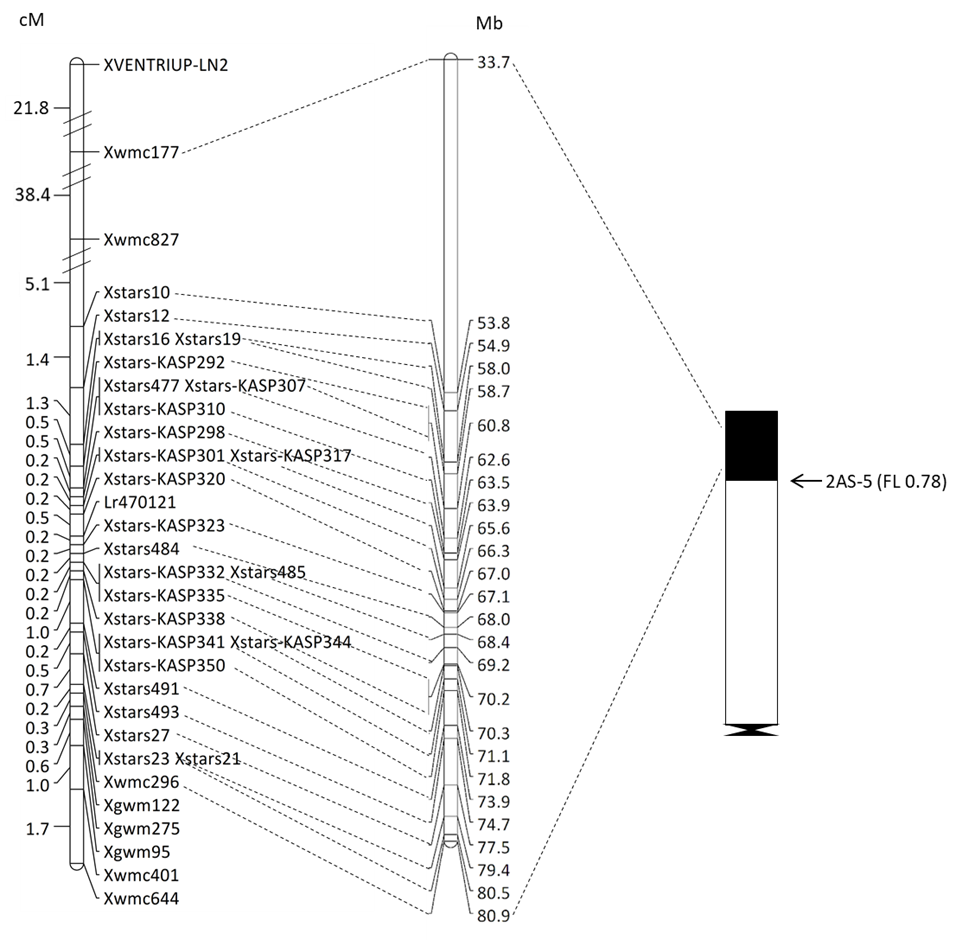
Figure 1. Linkage (left), physical (middle) and deletion bin map (right) of Lr81 (formerly Lr470121). Markers are shown on the right side of the linkage map with their genomic locations given in the physical map. Genetic distances in cM are presented on the left of the linkage map. Molecular markers flanking Lr81 were connected to their appropriate physical bin. The breakpoint of the Chinese Spring deletion line 2AS-5 is shown with an arrow, and the corresponding fraction length (FL) value is given in parenthesis.
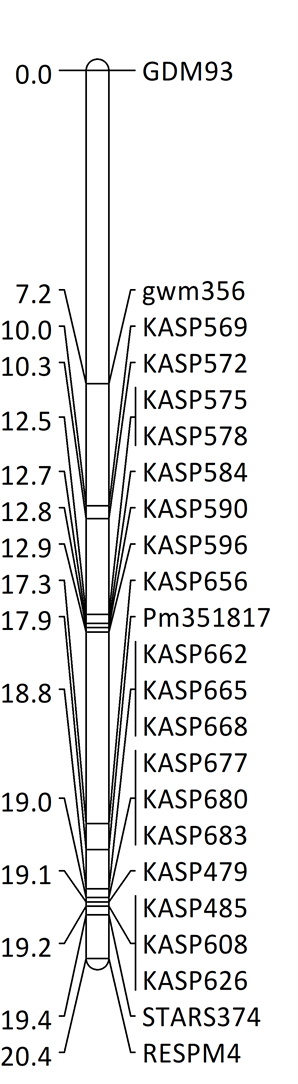
Figure 2. A linkage map containing Pm351817. Molecular markers are shown on the right side of the linkage map, and genetic distances in cM are on the left side.
| Source | State | Region | PI 351817 (Pm351817) | Xinmai 208 (Pm65) | Jagalene |
|---|---|---|---|---|---|
| GAP-A-2-3 | Georgia | Southwest | R | R | S |
| GAP-B-2-2 | Georgia | Southwest | R | R | S |
| MSG-C-3-4 | Georgia | Southwest | R | R | S |
| NCF-D-1-1 | North Carolina | Mid Atlantic | R | R | S |
| NCC-B-1-3 | North Carolina | Mid-Atlantic | R | R | S |
| NYA-E-3-3 | New York | Great Lakes | R | R | S |
| NYB-E-1-2 | New York | Great Lakes | R | S | S |
| OKH-A-2-3 | Oklahoma | Great Plains | R | R | S |
| OKS-A-2-2 | Oklahoma | Great Plains | R | R | S |
| OKS-B-2-2 | Oklahoma | Great Plains | R | R | S |
| PAF-E-2-2 | Pennsylvania | Great Lakes | R | R | S |
PAF(14)-D- 1-2 | Pennsylvania | Great Lakes | R | R | S |
MIR(14)- D-3-3 | Michigan | Great Lakes | R | R | S |
MIR(14)-E- 1-3 | Michigan | Great Lakes | R | R | S |
MTG1-3a | Montana | Northwest | R | R | S |
MTG1-1a | Montana | Northwest | R | R | S |
NEI 3-1 | Nebraska | Great Plains | R | R | S |
NEI 1-3 | Nebraska | Great Plains | R | R | S |
NEI 5-5 | Nebraska | Great Plains | R | R | S |
R and S represent resistant and susceptible responses, respectively.
Novel bird-cherry oat aphid and greenbug resistance sources
Bird cherry-oat aphid (BCOA) poses a persistent threat to wheat production in Oklahoma. The reported yield losses caused by BCOA can vary up to 85%, depending on the extent of the infestation, the plant growth stage at infestation and whether the aphids transmit barley yellow dwarf virus (BYDV) to wheat. Currently, few BCOA resistance sources are available for wheat improvement beyond the effort WIT devoted in the past five years to selecting intrinsically for BCOA tolerance in adapted breeding populations. WIT must remain vigilant in identifying new and potent BCOA resistance sources in more extended wheat gene pools.
WIT developed a new BCOA assay protocol and evaluated the responses of a set of 260 Ae. tauschii 260 lines which account for 48% of the total Ae. tauschii accessions available in global germplasm banks. A set of over 15 lines exhibited resistance. These lines have the potential to enhance wheat BCOA resistance, and a project aimed at developing BCOA-resistant synthetic hexaploid wheat has been initiated.
Greenbug [Schizaphis graminum (Rondani)] is also a major wheat pest in the U.S., and its periodic, widespread outbreaks that usually occur every 5-10 years in the Great Plains cause significant yield losses in this region. To date, eight greenbug resistance genes (Gb1- Gb8) have been officially designated with four originating from Ae. tauschii. Given that greenbug resistance genes usually confer resistance to only one or a few greenbug biotypes, and new biotypes continuously emerge in fields, identification of novel resistance sources is imperative for sustainable wheat production. Thus, WIT further evaluated the abovementioned Ae. tauschii panel for responses to greenbug biotypes C, E, F, G, I and TX1. A total of 102 lines exhibited resistance to at least one biotype with 14 different resistance profiles or phenotypes, indicating that Ae. tauschii is a valuable greenbug resistance source. Currently, a set of synthetic wheat lines derived from some of these greenbug-resistant Ae. tauschii lines are being evaluated, and crosses will be made to transfer novel greenbug resistance genes into elite Oklahoma breeding lines in 2023.
Pyramiding novel resistance genes
Transfer of resistance genes into WIT breeding lines continued in the 2021-2022 season via marker-assisted selection. These included leaf rust resistance genes Lr81, Lr622111, Lr47 and Qlr.stars-1RS; powdery mildew resistance genes Pm59, Pm63, Pm65 and Pm351817; and greenbug resistance gene Gb8.
Acknowledgements
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
