Adaptive Introgression 2023
Identification of novel QTLs for leaf rust resistance
Globally, about 21% of wheat production losses are due to pests and diseases. Leaf rust, caused by Puccinia triticina (Pt), is the leading cause. Yield losses from leaf rust are mainly due to reduction in kernel number and size, resulting in huge economic losses worldwide. Host resistance is the most efficient, cost-effective, and environment-friendly approach to managing leaf rust. Unfortunately, only a small fraction of the genetic diversity was sampled at each step of wheat evolution and domestication. Wheat breeding over the past two centuries – especially after the universal adoption of high-yielding dwarf cultivars during the “Green Revolution” – further created genetic diversity bottlenecks that led to the loss of many leaf rust resistance genes. Therefore, wheat landraces and relatives remain to be important sources for leaf rust resistance genes.
The Oklahoma State University Wheat Improvement Team (WIT) identified three quantitative trait loci (QTL) for leaf rust resistance in the Uzbekistani wheat landrace Teremai Bugdai, QLr-Stars-2DS, QLr-Stars-6BL, and QLr.stars-7B (Figure 1).
QLr-Stars-2DS, which is either a new Lr2 allele or a new resistance locus, was delimited to a 19.47 Mb interval between 46.4 Mb and 65.9 Mb on 2DS and explained 31.3% - 33.2% of the phenotypic variance in two experiments.
QLr-Stars-6BL was mapped in an 84.0 Kb interval between 719.48 Mb and 719.56 Mb on 6BL, accounting for 33% to 36.8% of the phenotypic variance.
QLr.stars-7BL was placed in a 350 Kb interval between 762.41 Mb and 762.76 Mb on 7BL and explained 4.4% to 5.3% of the phenotypic variance.
Nine kompetitive allele specific PCR (KASP) markers were developed for tagging these QTLs in breeding populations. As shown in Figure 2, leaf rust resistance can be significantly enhanced by combining these QTLs, and satisfactory selection results can be achieved when these KASP markers are used in marker-assisted selection.
Discovery and characterization of the novel greenbug resistance gene Gb
Greenbug (Schizaphis graminum) is a serious cereal insect pest worldwide. Feeding greenbugs inject saliva containing pectinesterase and polygalacturonase into plants to facilitate the extraction of plant sap. This deprives the cereal host of water and nutrients, leading to chlorotic and necrotic spots on leaves, plant death, and yield loss. In addition, greenbug is a vector of viruses, such as maize dwarf mosaic virus and barley yellow dwarf virus. Its feeding activity also predisposes plants to diseases to cause more severe damage to plants. The emergence of new greenbug biotypes in the field makes it urgent to identify novel greenbug resistance genes in wheat.
WIT identified a new greenbug resistance gene, officially designated as Gb9, in the synthetic hexaploid wheat (SHW) line CWI 76364 (PI 703397). Genetic analysis delimited Gb9 to a 0.6-cM interval flanked by KASP markers located at 599,835,668 bp (Stars-KASP872) and 600,471,081 bp (Stars-KASP881) on 7DL (Figure 3). Gb9 was 0.5 cM distal to Stars-KASP872 and 0.5 cM proximal to Stars-KASP881. Allelism tests indicated that Gb9 is a new greenbug resistance gene that confers resistance to greenbug biotypes C, E, H, I, and TX1 (Table 1).
TX1 is one of the most widely virulent biotypes and has overcome most known wheat greenbug resistance genes. The introgression of Gb9 into locally adapted wheat cultivars is of economic importance, and the KASP markers developed in this study can be used to tag Gb9 in cultivar development.
Identification of new stripe rust resistance sources
Stripe rust caused by Puccinia striiformis f. sp. tritici (Pst) is an economically important wheat disease worldwide with annual losses of over $1 billion. Stripe rust epidemics occur frequently in major wheat-producing countries, such as the USA, UK, China, and Australia, etc. (two or three years every five years). New stripe rust resistance sources are needed for developing stripe rust-resistant cultivars. WIT tested a set of wheat accessions using five current U.S. Pst races PSTv-4, PSTv-14, PSTv-37, PSTv-40, and PSTv-52, leading to the identification of 12 resistant accessions. Of these, one historical cultivar, three landraces, and two synthetic hexaploid wheat accessions were resistant to all five Pst races
Currently, WIT is genotyping and phenotyping recombinant inbred line populations from crosses PI 622129 × TAM 110 and PI 62004 × Jagalene to identify the stripe rust resistance genes in PI 622129 and PI 62004. Notably, both PI 622129 and PI 62004 exhibited a wide spectrum of resistance to leaf rust with low infection types to all 19 current U.S. Pt races used in our studies. Characterization and introgression of both leaf and stripe rust resistance genes in these accessions are of great importance for sustainable wheat production in the U.S. pyramiding novel resistance genes.
WIT continued to transfer newly identified resistance genes into WIT breeding lines in the 2022-2023 season via marker-assisted selection. These included leaf rust resistance genes Lr81, Lr622111, Qlr.stars-1RS, QLr-Stars-2DS, and QLr-Stars-6BL; powdery mildew resistance genes Pm59, Pm63, Pm65, and Pm351817; and greenbug resistance genes Gb8 and Gb9.
Acknowledgments
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
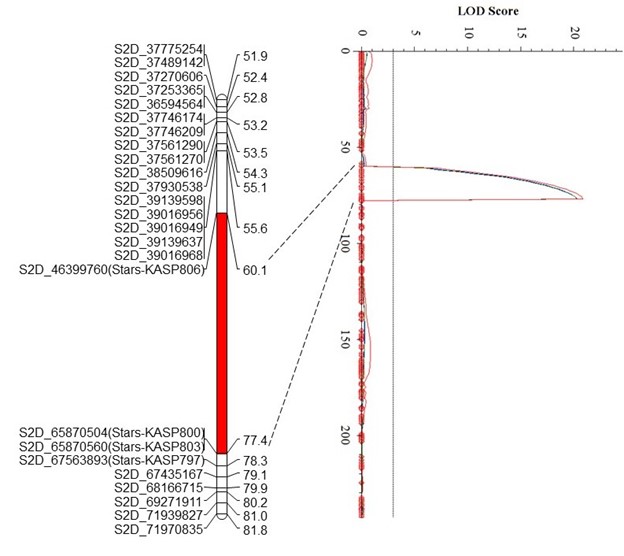
Figure 1A
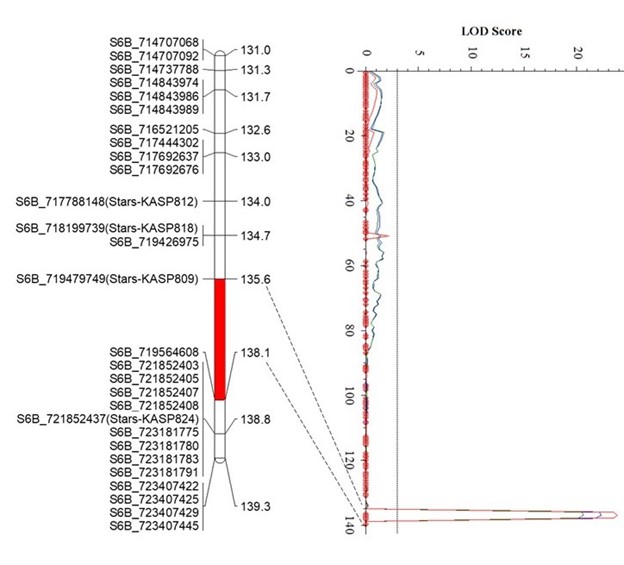
Figure 1B
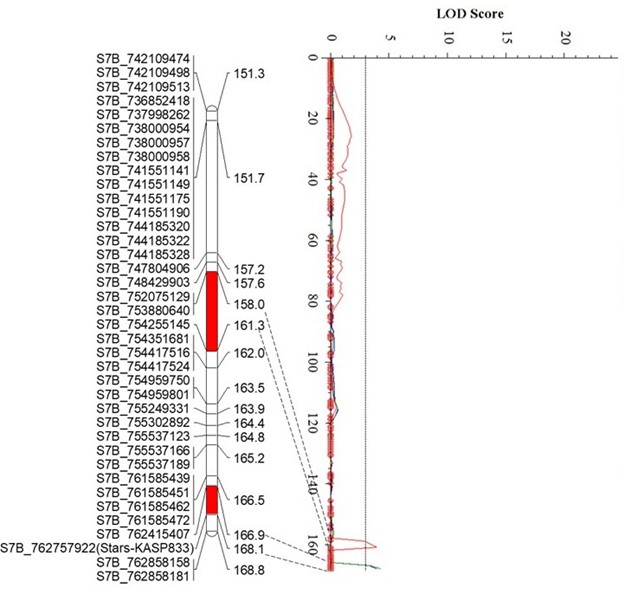
Figure 1. Figure 1. Partial linkage maps showing locations of QLR-Stars-2DS (A), QLR-Stars-6BL (B), and QLR-Stars-7BL (C) on their corresponding chromosomes. Red, green, and blue represent Experiment 1, Experiment 2, and the mean data of two experiments, respectively. GBS-SNP, represented by the genomic location in IWGSC RefSeq v2.1, and KASP markers are shown on the left. Genetic distances are shown on the right of the linkage maps. The vertical dot lines mark the threshold logarithm-of-the-odds value of 3.0, and the confidence locations of QTLs are highlighted in red bars on the maps.
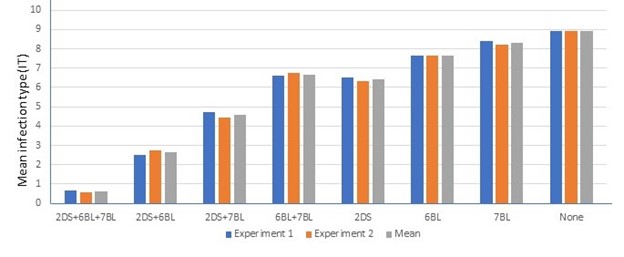
Figure 2. Mean infection types (IT) of recombinant inbred lines (RILs) with or without QLR-Stars-2DS (2DS), QLR-Stars-6BL (6BL), and QLR-Stars-7BL (7BL) in Experiment 1 (blue), Experiment 2 (orange), and their average (gray). A 0-9 scale was used to score the infection type (IT) of each plant, and RILs were classified as highly resistant (IT = 0-3), moderately resistant (IT = 4-5), moderately susceptible (IT = 6-7), and highly susceptible (IT = 8-9), respectively.
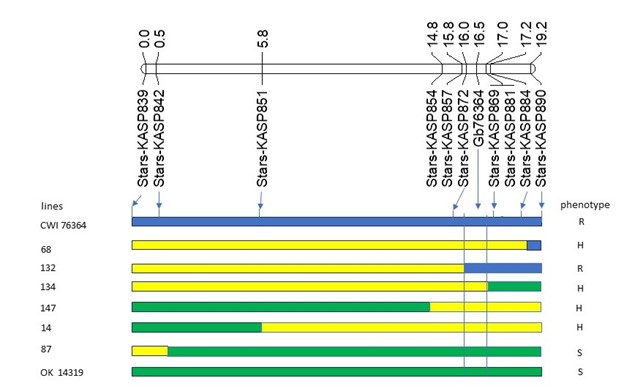
Figure 3. Linkage map showing genomic location of Gb9 (formerly Gb76364), graphic genotypes and phenotypes of CWI 76364, OK 14319, and a few F2:3 lines. KASP markers and genetic distance in cM were given below and above the linkage map, respectively. Blue, green, and yellow represent CWI 76364, OK 14319, and heterozygous genotypes, respectively. R, resistant; S, susceptible; H, heterozygous.
| Wheat line (gene) | B | C | H | I | FL | TX1 |
|---|---|---|---|---|---|---|
| CWI 76364 (Gb9) | S | R | R | R | S | R |
| Largo (Gb3) | S | R | R | R | S | S |
| CI 17959 (Gb4) | S | R | S | R | S | S |
| W7984 (Gb7) | S | S | S | R | S | S |
| PI 697274 (Gb8) | R | R | R | R | R | S |
| Jagalene | S | S | S | S | S | S |
