Adaptive Introgression
Insects and diseases cause most of the wheat yield losses globally. This component of the Wheat Improvement Team (WIT) is dedicated to using multiple tools from several disciplines, including wheat pathology, entomology, molecular genetics and wheat pre-breeding to diversify and fortify the germplasm base of the WIT variety development pipeline.
Novel Leaf Rust Resistance Genes
Leaf rust, caused by Puccininia triticina (Pt), is one of the most common wheat diseases in the United States Great Plains, where new Pt races frequently occur. Currently, most leaf rust resistance genes have lost effectiveness in this region, and new genes and gene packages are needed to sustain wheat production. Gene Lr47, originally identified on the wheat-T. speltoides 7A.7S translocation segment 7S#1 in accession CI 17884, is one of only a few race-specific resistance genes still effective in the Great Plains. A shorter interstitial translocation chromosome segment carrying Lr47, 7S#1S was previously generated from 7S#1 in the absence of the Ph1b gene and transferred to spring wheat cultivar Pavon F76 using backcrossing, which led to the development of PI 603918. To determine the genomic location of Lr47, a set of 7A-specific simple sequence repeat (SSR) markers was developed to genotype CI 17884, Pavon F76, a pair of near-isogenic lines differing in the presence or absence of 7S#1S and PI 603918. Haplotype analysis indicated the length of the 7S#1S translocation was 157.23 to 174.42 Mb, accounting for about 23% of the 7A chromosome.
Further quantitative trait locus analysis identified an incomplete gene for leaf rust resistance, designated QLr.stars-1RS, on the 1BL.1RS translocation segment in Bainong 418 (Figure 1). Bainong 418 is a high-yielding cultivar widely grown in the Huang-Huai River Valleys of China. Bainong 418 exhibits a mesothetic infection type with various pustule sizes and hypersensitive flecks, which is consistent with partial resistance expression. A population of recombinant inbred lines (RILs) from CI 17884 x Bainong 418 was genotyped using single nucleotide polymorphisms (SNP) markers. Phenotypic analysis indicated the combination of Lr47 and QLr.stars-1RS significantly enhanced leaf rust resistance (Figure 2). To facilitate rapid introgression of Lr47 and QLr.stars-1RS into Oklahoma wheat cultivars, two SNPs on 7S#1S and four SNPs on chromosome arm 1RS were converted to kompetitive allele specific PCR (KASP) markers. Of these, one and two KASP markers are specific to the 1RS chromosome arm (KASP-1RS-1) and the 7S#1S translocation segment (KASP-Lr47-1 and KASP-L47-2), respectively. These markers are effective when used to select Lr47 and QLr.stars-1RS in breeding populations.
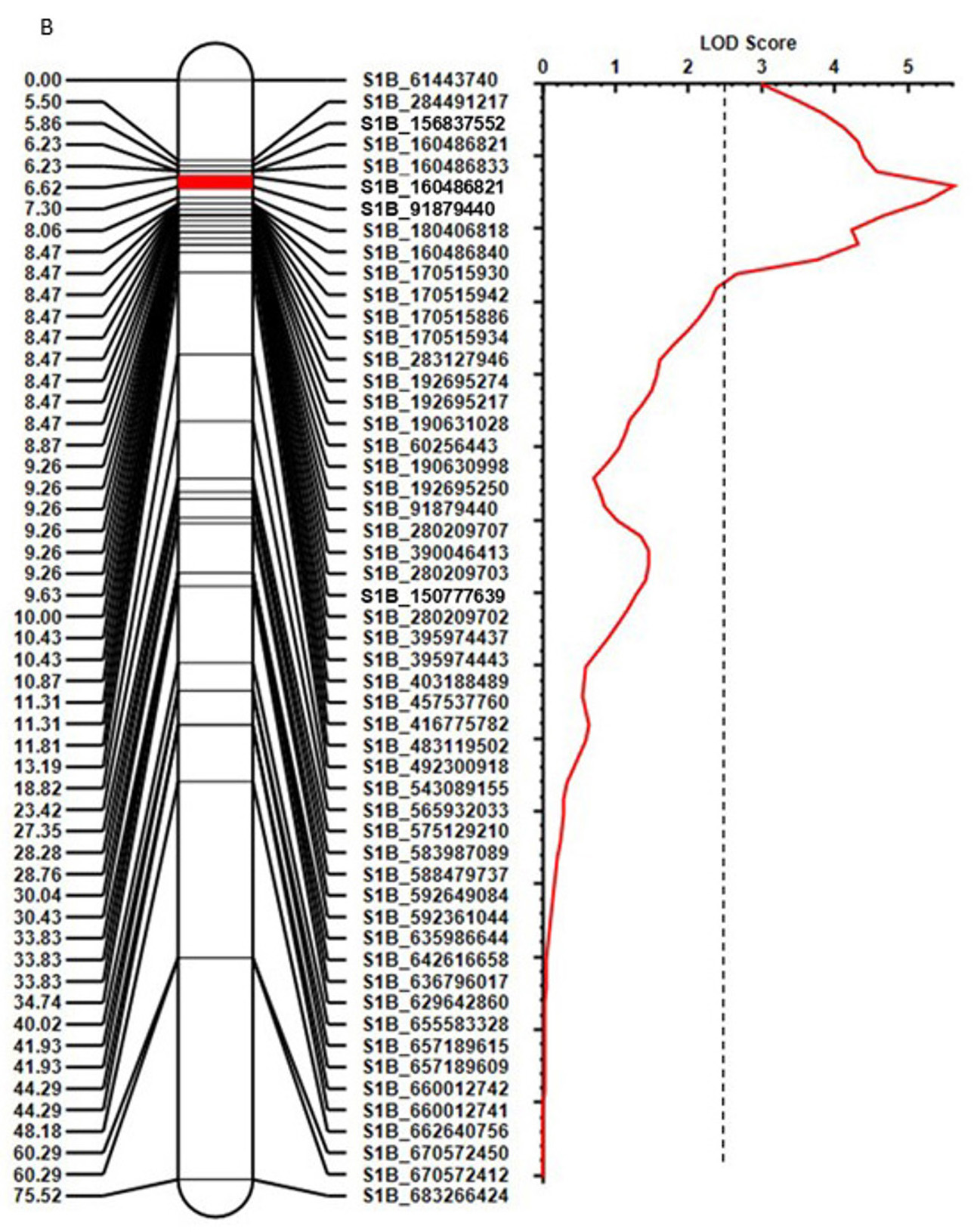
Figure 1. Composite linkage map of the incomplete leaf rust resistance gene QLr.stars-1RS. Genetic distance (in CM) is shown on the left, and the single nucleotide polymorphisms derived from genotyping-by-sequencing, represented by the genomic location in the IWGSC reference RefSeq v.1.0, are shown on the right of the linkage map. The dotted vertical rule marks the threshold log of odds value of 2.5. The confidence location of QLr.stars-1RS is highlighted in red.
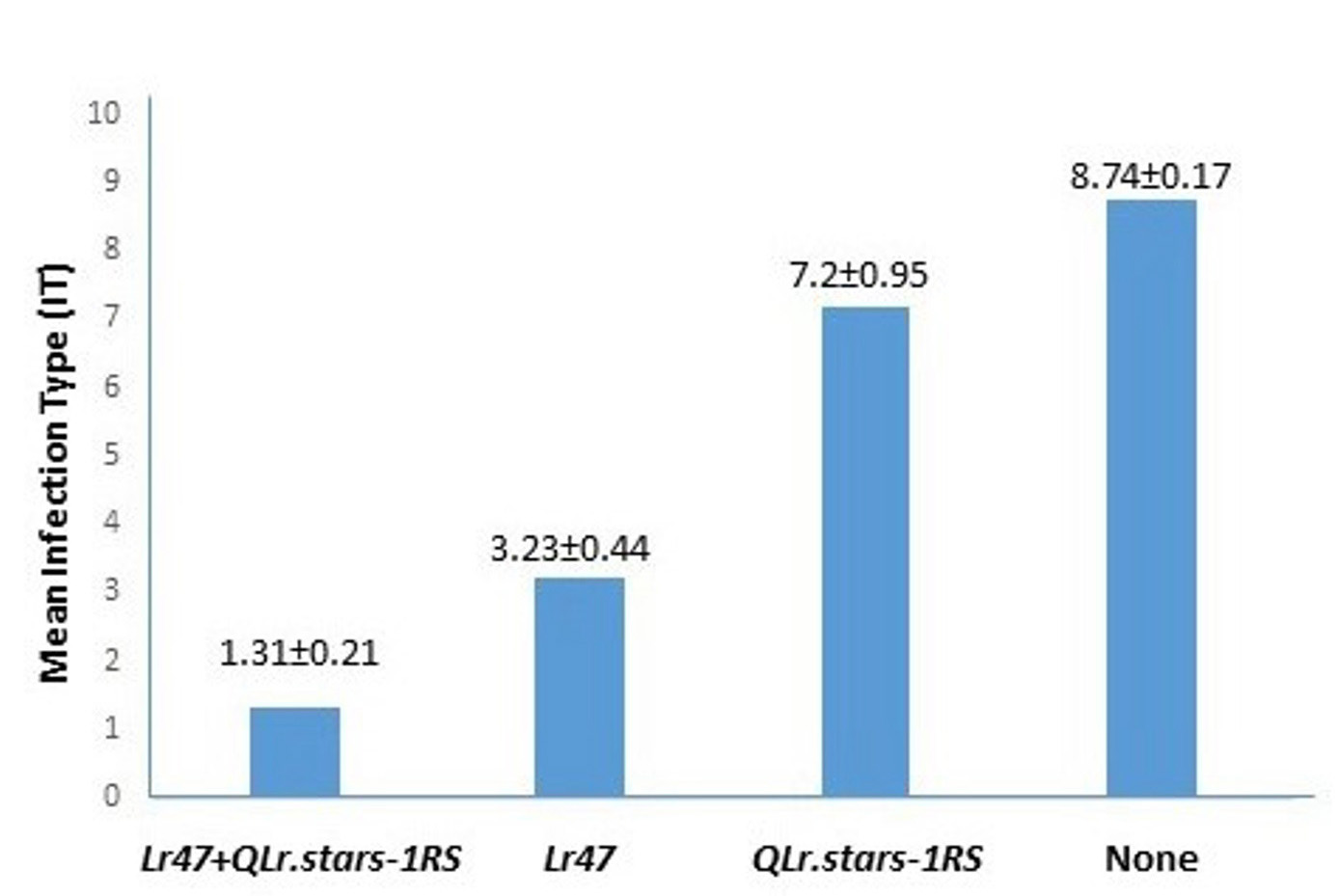
Figure 2. Mean infection types of recombinant inbred lines (RILs) with or without Lr47 and QLr.stars-1RS. The 95% confidence interval of infection type (IT) value for each group of RILs is given on the top of the corresponding bar. A zero-to-nine scale was used to determine IT of each RIL, with zero the most resistant and nine the most susceptible.
Additionally, the highly effective leaf rust resistance gene Lr622111 was reported in 2019. The Wheat Improvement Team also developed kompetitive allele specific PCR (KASP) markers linked to Lr622111 to transfer this gene into Oklahoma wheat cultivars via marker-assisted backcrossing.
Novel Greenbug Resistance Genes
Thirty biotypes of greenbugs [Schizaphis graminum (Rondani)] have been identified in the U.S. Each biotype is virulent to a few wheat greenbug resistance genes. Gene Gb1 confers resistance to some important biotypes and is the only gene conferring resistance to biotype F. Therefore, the combination of Gb1 with other genes, such as Gb3 and Gb8, is one way to develop durable greenbug-resistant cultivars. The recessive gene action of Gb1 makes the use of DNA markers a more efficient breeding strategy. With the support of the Oklahoma Wheat Research Foundation, Gb1 was mapped using F2 and F2:3 populations derived from the cross DS28A/Custer. A bulked segregate analysis revealed SNPs closely associated with Gb1 on the short arm of chromosome 1A in DS28A. Linkage analysis based on SSR markers assigned Gb1 to an interval flanked by molecular markers WMC818 and WMC24. Future expectations are to fine-map Gb1 and develop KASP markers for stage-one introgression into Oklahoma cultivars in 2021.
Introgression Summary
In previous reports, WIT discovered a few biotic stress resistance genes, including leaf rust resistance genes Lr470121 and Lr622111; powdery mildew resistance genes Pm59, Pm63 and Pm65; and greenbug resistance gene Gb8. WIT continued to introgress these genes into Oklahoma wheat cultivars via marker-assisted backcrossing in 2020, and new projects aimed at transferring Lr47, Gb1, Gb5 and QLr.stars-1RS into OSU breeding lines were initiated.
