Gene Discovery & Genomic Application
Unique alleles of Lr34 in Southern Plains varieties
The wheat resistance gene Lr34, or the locus representing Lr34/Yr18/Sr57/Pm38, confers non-specific resistance to fungal diseases, including leaf rust, stripe rust, stem rust and powdery mildew, and to the viral disease, barley yellow dwarf. Because of the durability and non-specificity of its resistance against multiple pathogens, Lr34 has become one of the most important disease-resistant genes in wheat worldwide. Smith’s Gold and Iba are two Oklahoma State University varieties that contain Lr34 but not the Lr34 allele present in Duster.
Allelic variation in Lr34 was revealed to rely on functional polymorphisms in exon 11 and exon 12 when this gene was initially cloned and reported in the journal Science in 2009. In previous research from this lab, polymorphisms in exons 11 and 12 did not align with resistance differences between two winter wheat varieties: 2174, carrying the resistance allele Lr34a, and Jagger, carrying the susceptible allele Lr34b. The Lr34b allele has a novel mutation in exon 22, which causes a premature stop codon of Lr34b, resulting in a non-functional protein product lacking 185 amino acids. The susceptible Lr34b allele is ordinarily selected out in Wheat Improvement Team (WIT) breeding populations.
In a study recently published from this lab, another novel allele in the OSU winter wheat variety Duster was discovered. The global wheat research community believes Lr34 exists as one copy on the short arm of chromosome 7D in all hexaploid wheat varieties, but current dogma does not apply to Duster. WIT found Duster has both alleles Lr34a and Lr34b, which is thus referred to as Lr34ab. This coexistence of two genes in Duster is due to a duplication of a chromosomal fragment that includes Lr34 (Figure 1). With partial support from the Oklahoma Wheat Research Foundation, the function of the Duster Lr34ab allele was corroborated by leaf-tip necrosis and leaf rust resistance. This newfound duplicated event not only challenges the current dogma that Lr34 exists as one copy, but also conveys caution to breeders using conventional markers to track Lr34 in breeding populations. New diagnostic kompetitive allele-specific PCR (KASP) markers and protocols were developed for multiple alleles of Lr34, already extensively utilized by WIT, and eventually, in the global wheat community.
KASP Markers for QYld.osu-1BS, a grain yield QTL
In the previous year of this project, a grain yield gene was demonstrated in Duster at the QYld.osu-1BS locus on chromosome arm 1BS. Among 6,820 single nucleotide polymorphisms (SNPs) in the 25.3 Mb QYld.osu-1BS region, Duster had 1,102 bp of unique SNPs compared with Billings and Chinese Spring. In comparison, Billings had 432 bp of unique SNPs compared with Duster and Chinese Spring.
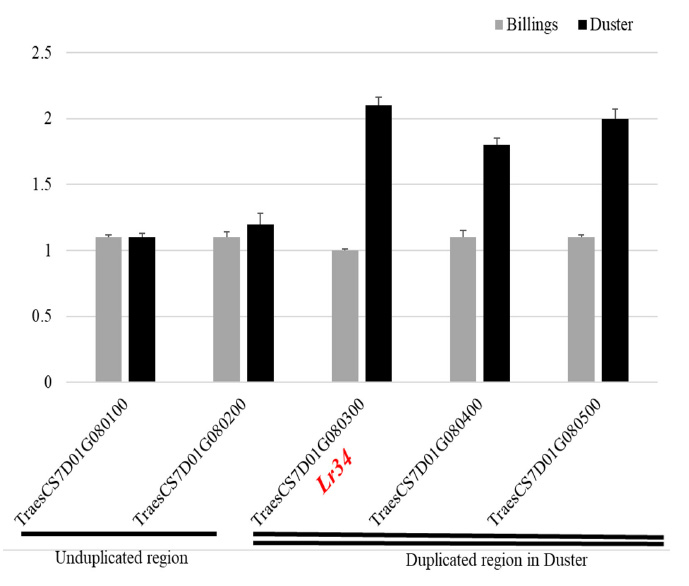
Figure 1. A chromosomal fragment duplication event in Duster but not in Billings. qRT-PCRs were used to determine copy number of five genes, including Lr34, two neighboring genes on its distal side (TraesCS7D01G080100 and TraesCS7D01G080200) and two neighboring genes on its proximal side (TraesCS7D01G080400 and TraesCS7D01G080500). The single line under TraesCS7D01G080100 and TraesCS7D01G080200 indicates the two genes have one copy in both Billings and Duster. The double lines under TraesCS7D01G080300, TraesCS7D01G080400 and TraesCS7D01G080200 indicate that each of these three genes has two copies in Duster (black bar) but one copy in Billings (gray bar).
PCR markers were developed for the unique sequence in Duster, and these markers were used to screen up to 135 accessions, including 100 hexaploid wheat varieties and 35 wheat relatives. Only five accessions (PI 278581, PI 352324, PI 352326, PI 542273 and PI 599155) showed the same allele as Duster. However, these five accessions all are diploid or tetraploid wheat among the 100 hexaploid wheat varieties, including 57 released varieties and breeding lines used in the U.S. Wheat Coordinated Agricultural Project (CAP). Duster was the only wheat variety with QYld.osu-1BS.
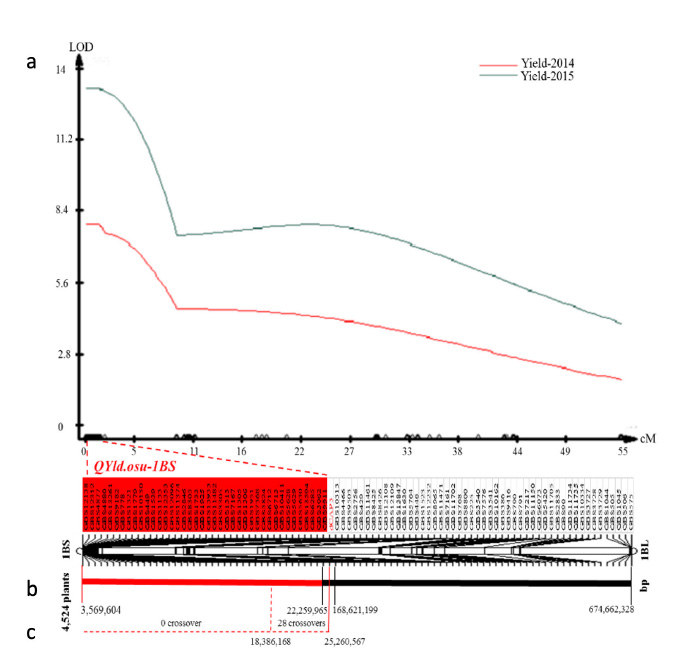
Figure 2. Genetic mapping and physical location of QYld.osu-1BS. (a) QYld.osu-1BS was mapped to the distal region of chromosome arm 1BS. Yield-2014 and Yield-2015 indicate grain yields measured in the field in 2014 and 2015, respectively. The QTL was declared since the logarithm of the odds (LOD) score exceeded the threshold of 2.5 (the dotted line). Forty genotyping by sequencing markers that are clustered as a group and show no crossovers in the Buster recombinant inbred line (mapping) population are shaded in red. The dCAP PCR marker dCAP3 is highlighted in red. B) The physical location of QYld.osu-1BS. QYld.osu-1BS was initially mapped to cover the telomere to 168,621,199 bp but narrowed to 25,260,567 bp by dCAP3. C) Locations of 28 crossovers in the mapping population. All 28 recombinant events were found in the region between 18.4 Mb and 25.3 Mb.
The unique sequences in Duster were converted into high-throughput KASP assays. KASP-12 was designed for the SNP at 1,253,260 bp, KASP-17 was designed for the SNP at 10,104,175 bp, and KASP-8 was designed for the SNP at 18,386,168 bp. These three unique markers representing the Duster allele spanning QYld.osu-1BS will be used to ensure the entire genomic region of the Duster allele at QYld.osu-1BS is preserved in WIT breeding lines derived from Duster.
A Major Gene for Spikelet Node Number and Grain Yield in Common Wheat
From OSU’s participation in the U.S. Wheat CAP, a major quantitative trait locus (QTL) that governs spikelet node number was discovered in two spring wheat varieties (Figure 3a). The candidate gene was mapped on chromosome arm 7BL and cloned using a map-based cloning approach. Transgenic wheat plants were successfully generated in this lab that may allow WIT to show the function of the candidate gene for spikelet number per spike in wheat under photoperiod and temperature-controlled greenhouse conditions. The favorable allele of the targeted QTL was cloned into a spring wheat variety.
Subsequently, Billings was found to carry the favorable allele (Figure 3b). Among 50 U.S. wheat varieties, Billings is one of only three varieties that carry this yield-enhancing allele. Current research in this project is committed to testing its effects in various winter wheat backgrounds.

Figure 3a.

Figure 3b.
Figure 3. Phenotypes of transgenic wheat plants carrying a candidate gene for spikelet node number. a) Comparative phenotypes for arbitrary A and B alleles used to clone the gene for spikelet node number and b) Comparative phenotypes of Billings and Duster grown in a greenhouse.
