Gene Discovery Transformation & Genomic Applications
The ultimate goal of public pre-breeding programs is to identify ideal genotypes and develop molecular tools that can be used to accelerate breeding of new varieties with increased grain yield. In our recent studies, we have reached a milestone in groundbreaking discoveries of a gene called TaCol-B5 that could potentially lead to increased grain yield in common wheat.
Wheat grain yield is influenced by three components: the number of spikes per unit land area, the number of kernels per spike and kernel weight. The final number of kernels per spike is influenced by two subcomponents: spikelet nodes per spike and the kernel number per spikelet. Therefore, a better understanding of the genetic control of these components and subcomponents would help in identifying new approaches to increase wheat grain yield without adding more fertilizers or other agricultural inputs. Wheat grain yield can be increased by increasing any single part of these components and subcomponents while the others remain unchanged, but there are trade-off relationships among the components and subcomponents. For example, trade-offs exist between spike number and kernel number per spike and between kernel number per spike and kernel weight. The trade-off between three individual yield components is a major bottleneck for further yield improvement. A strategy can be utilized to genetically counteract the trade-off relationships for increases of yield potential.
Wheat has a unique pattern of spike architecture compared with other crops. A normally developed spike in contemporary wheat cultivars can generate 16 to 25 spikelet nodes, and almost all spikelets are fully fertile with the possible exception of spikelets on the upper and lower portions of a spike, which may generate fewer kernels and could even be sterile. A normal spikelet in contemporary wheat cultivars produces 5 to 12 florets contained by two small glumes, but only 1 to 5 of the differentiated florets may set seed, whereas more than 70% of florets are aborted during development. Within the same spikelet, kernels at the first and second positions are much larger than those at the third, fourth or higher positions. The higher the position at which a kernel resides in the spikelet, the less it will weigh, pointing to a trade-off between kernels per spikelet and kernel weight. Understanding the developmental pattern of spikelets within a spike provides the opportunity to genetically increase the number of spikelet nodes, thereby increasing kernel number without decreasing average kernel weight.
WIT reported in previous Oklahoma Wheat Research Foundation annual reports that we mapped a major quantitative trait locus (QTL) for spikelet number on the long arm of chromosome 7B, called QSns.osu-7B. This QTL accounted for over 40% of the total observed or phenotypic variation. We cloned the gene responsible for this QTL, and named it TaCol-B5 to be more consistent with its orthologue (CONSTANS-like 5) in other plant species. When constitutively overexpressed in transgenic wheat tested in temperature-photoperiod controlled greenhouse conditions, TaCol-B5 was found to promote tillering and generate more spikelet nodes per spike, thereby increasing total spike number and seed number. When the transgenic wheat was grown under field conditions, TaCol-B5 still increased both traits. The transgenic wheat also showed extended spike length and decreased spikelet density, thereby modifying spike architecture (Figure 1). Ultimately, TaCol-B5 enhanced field-based grain yield of transgenic wheat by 11.9%. We published this work in Science on April 7, 2022, “The discovery of TaCol-B5 is a milestone toward enhancing yield in cereals because it improves our understanding of molecular mechanisms that control yield-related architectural traits” and “the identification of TaCol-B5 offers a new route to maximize yield in wheat,” according to Dr. Wilma van Esse, who wrote the perspective paper for Science.
It is noteworthy that the previously defined trade-off relationships could be broken down using TaCol-B5 that was over-expressed in transgenic wheat. In transgenic field and greenhouse experiments, TaCol-B5 increased total spike number while not decreasing kernel number per spike and increased kernel number while not decreasing kernel number. More kernels produced by TaCol-B5 will be located in the first and second floret positions in additional spikelets, whereas more kernels gained in the absence of increased spikelet numbers must set in the third, fourth or higher seed positions. TaCol-B5 can be used to remove some of this trade-off by extending the size of the “sink” to accommodate more spikes and spikelets and thus improve productivity under allowable environmental conditions.
We developed a diagnostic marker for the single nucleotide polymorphism (SNP), or single DNA sequence variant, involving the Ser269/Gly269 substitution between TaCol-B5 encoded by the dominant allele for higher yield and Tacol-B5 encoded by the recessive allele. Primers TaCOL-B5F1 and TaCOL-B5R1 were used to amplify an approximate 543 bp fragment. The polymerase chain reaction (PCR) products were digested with Pfl-MI restriction enzyme for overnight at 37°C and resolved on a 2% agarose gel. Polymorphic fragments comprised 543 bp from the TaCol-B5 allele and 302 bp + 241 bp from the Tacol-B5 allele (Figure 2). The Ser269/Gly269 substitution in TaCol-B5 and Tacol-B5 resulted in differential protein phosphorylation by TaK4 (Figure 3).
We used the diagnostic marker for TaCol-B5 to genotype 1,657 accessions in a global collection of modern hexaploid wheat cultivars and germplasm and found that only 2% of the international modern wheat panel possessed the same TaCol-B5 allele as the original source, cultivar CItr17600 from the International Maize and Wheat Improvement Center (CIMMYT). Distribution of the favored TaCol-B5 allele is illustrated in Figure 4. Among 49 U.S. wheat accessions, only three cultivars – Billings, McNeal and RSI5 (PI584453) – were found to have the TaCol-B5 allele. Among 57 U.S. hard red winter cultivars from the Great Plains, only the OSU’s Baker’s Ann was found to have the TaCol-B5 allele. We have extensively utilized Billings and Baker’s Ann and their descendants as parental lines in the OSU Wheat Improvement Team breeding populations.
*Note: Polymerase chain reaction refers to a laboratory technique for rapidly producing millions to billions of copies of a specific segment of DNA, which can then be studied in greater detail.
KASP Markers for Mutants of ACCase Genes
We modified Kompetitive Allele specific (KASP) markers for each of ACCase-A, ACCase-B and ACCase-D genes in contemporary wheat cultivars (Figure 5). Original KASP assays developed for detecting the ACCase mutations by Colorado State University involved PCR amplification of each of the three homoeologous ACCase genes, and the markers showed the expected genotype for ACCase-A and ACCase-D genes but not for the ACCase-B gene. The marker for the ACCase-B gene identified wild-type alleles in Smith’s Gold as expected but produced an inappropriate heterozygous reaction for three other varieties (Crescent AX, LCS-Fusion AX and LCS-Photon AX) possessing mutations for ACCase-A and ACCase-D genes. As we pooled leaf samples from at least three different plants, the original hypothesis was that heterogeneity existed at this locus within these three varieties for ACCase-B. However, when we tested six individual leaf samples from another CoAXium-confirmed cultivar, it was clear that the ACCase-B marker was not specific in the background of other ACCase mutants and produced false positive results. We concluded that the presence of SNPs in the primer binding sequences may suggest the appearance of genetic segregation when it truly does not exist. We optimized the reaction with re-combining different temperatures or obtaining mutant sequences for designing new KASP markers to resolve this issue. Our modified markers for each of three homoeologous ACCase genes have been used to genotype breeding materials provided by Dr. Brett Carver (see his report). The CoAXium component of the OSU wheat improvement program featured two seed-increase nurseries in 2022 and that material will be tested statewide in 2023 to arrive at effective mutations for herbicide tolerance in wheat.

Figure 1. Performance of TaCol-B5 overexpressed T2 transgenic plants in the field.
The image was produced to show significant differences between overexpressed TaCol-B5 (OE, transgenic plant #34) in Yangmai18 as a host plant and wild-type Yangmai18. The transgenic plants produced the longer and larger spikes than the wild-type. TaCol-B5-OE34 in transgenic Yangmai18 is pictured on the left while wild-type Yangmail18 is pictured on the right.
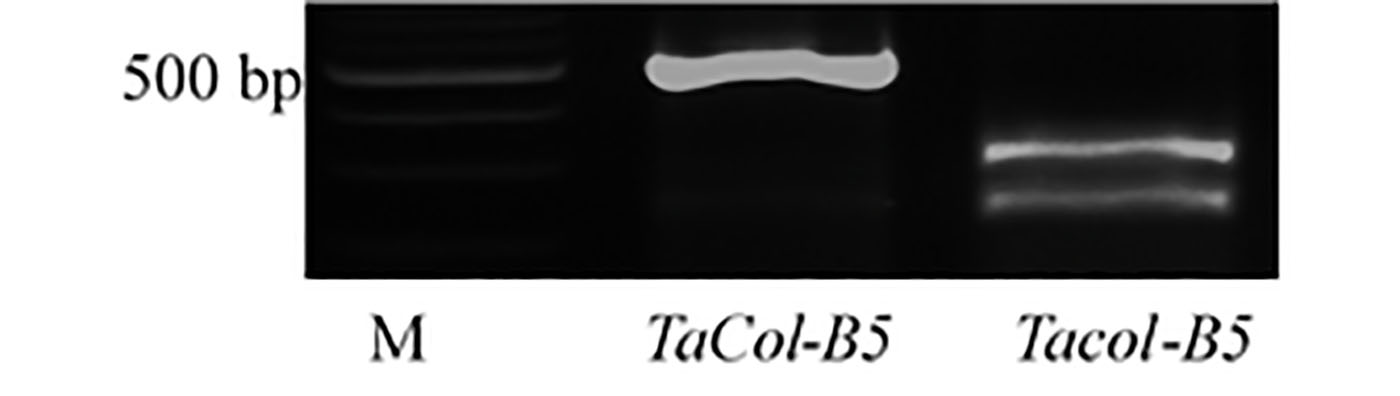
Figure 2. A gene marker for the SNP involving the Ser269/Gly269 substitution between TaCol-B5 and Tacol-B5.
The two primers, TaCOL-B5F1 and TaCOL-B5R1, amplified an approximate 543 bp fragment using this PCR program: 94°C for 3 mins, 40 cycles of 94°C for 30s, 60°C for 30s, and 72°C for 30s, following 72°C for 5 mins. 5% dimethyl sulfoxide (DMSO) was used in the PCR reaction. The PCR products were digested with Pfl-MI restriction enzyme overnight at 37°C and resolved on a 2% agarose gel. Polymorphic fragments were 543 bp from the TaCol-B5 allele in CItr 17600 and 302 bp + 241 bp from the Tacol-B5 allele in Yangmai18. M indicates DNA marker for molecular weight.
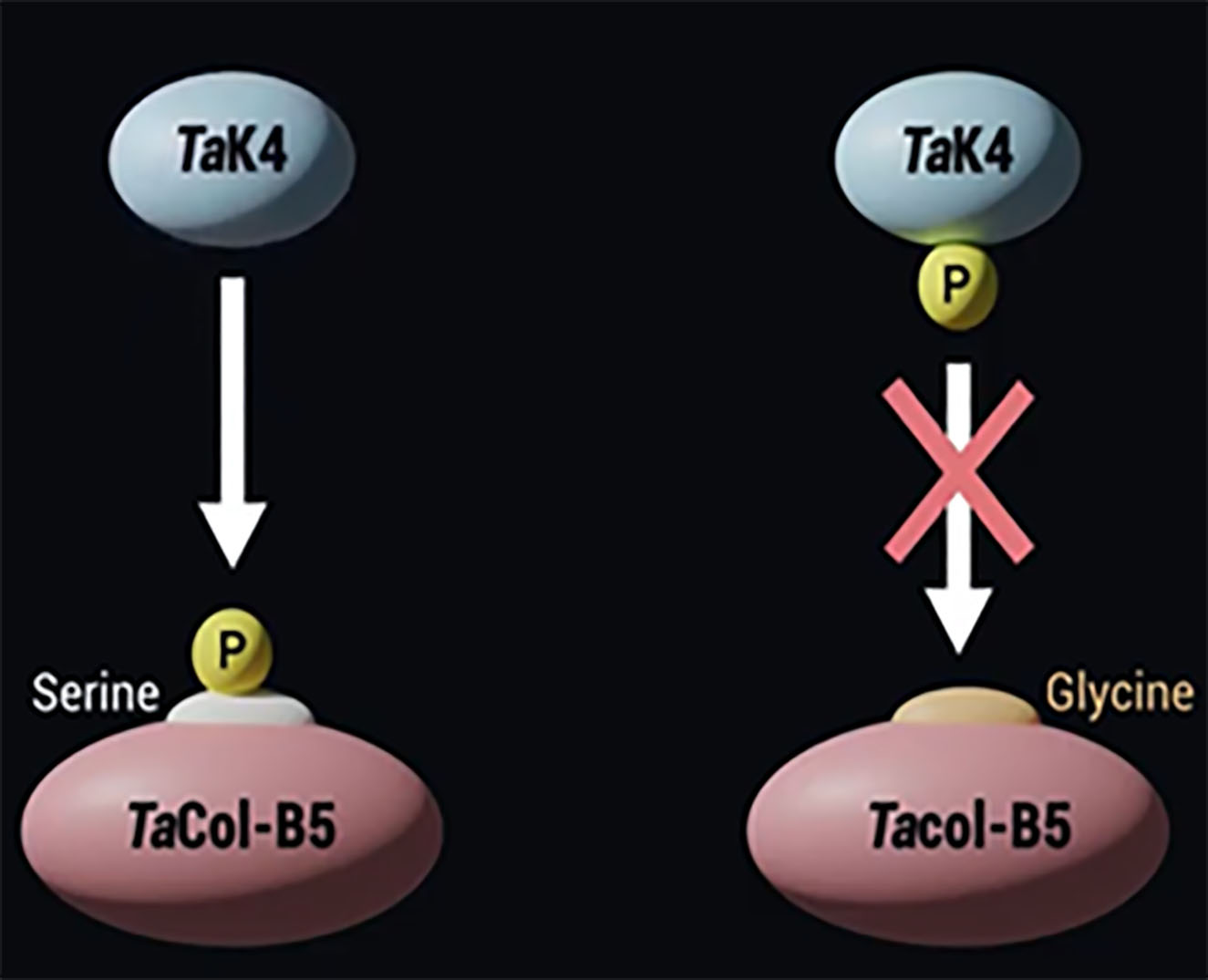
Figure 3. The differential phosphorylation of TaCOL-B5 by TaK4.
Amino acid serine in the position 269 of TaCol-B5 can be phosphorylated by TaK4, while amino acid glycine in the same position of Tacol-B5 cannot be phosphorylated by TaK4. The SNP involving the Ser269/Gly269 substitution between TaCol-B5 and Tacol-B5 was used to develop a diagnostic marker that differentiates the TaCol-B5 and Tacol-B5 alleles at the DNA level for molecular selection in breeding populations.
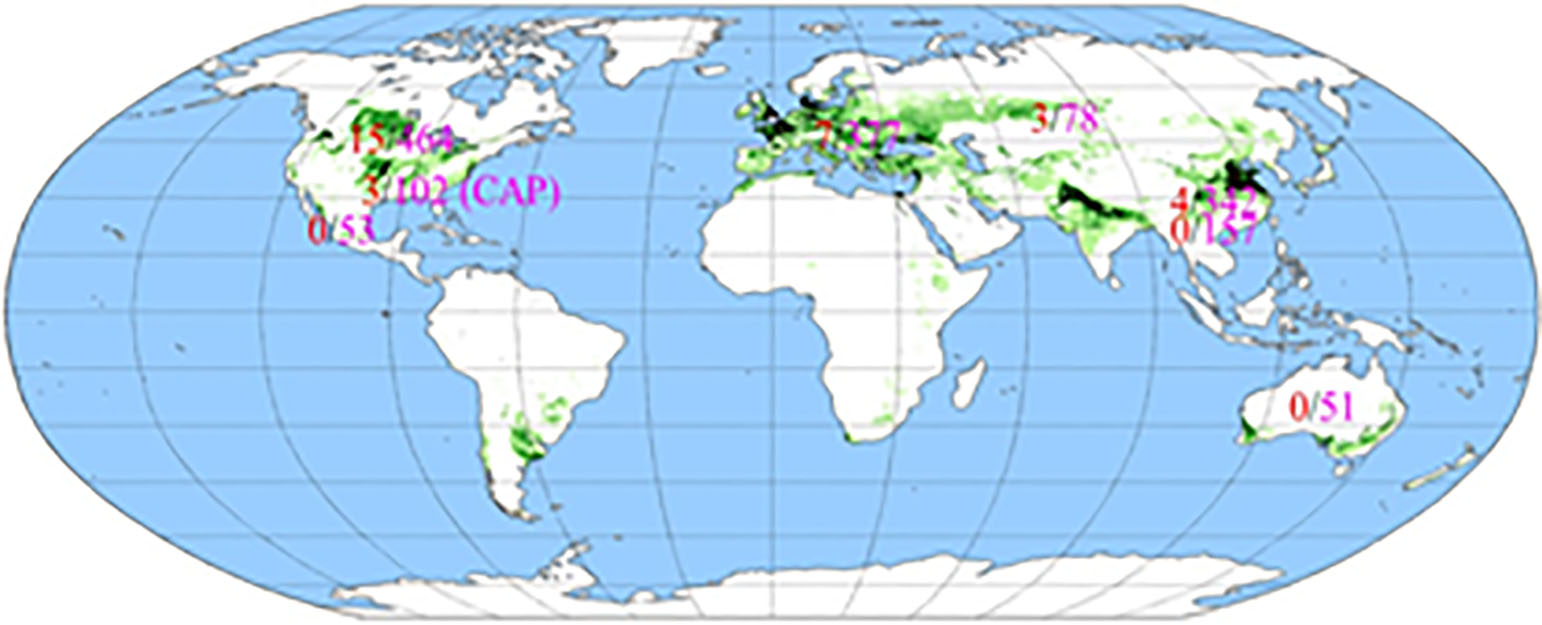
Figure 4. The global distribution of TaCol-B5 and Tacol-B5 alleles.
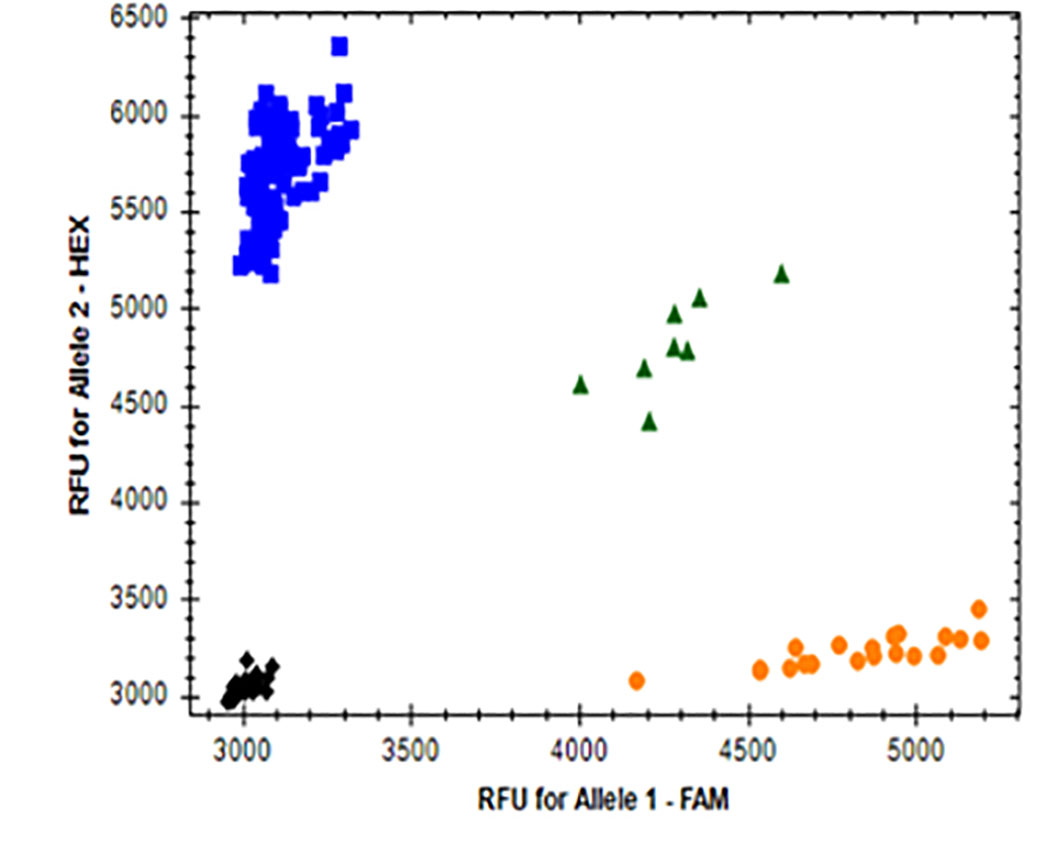
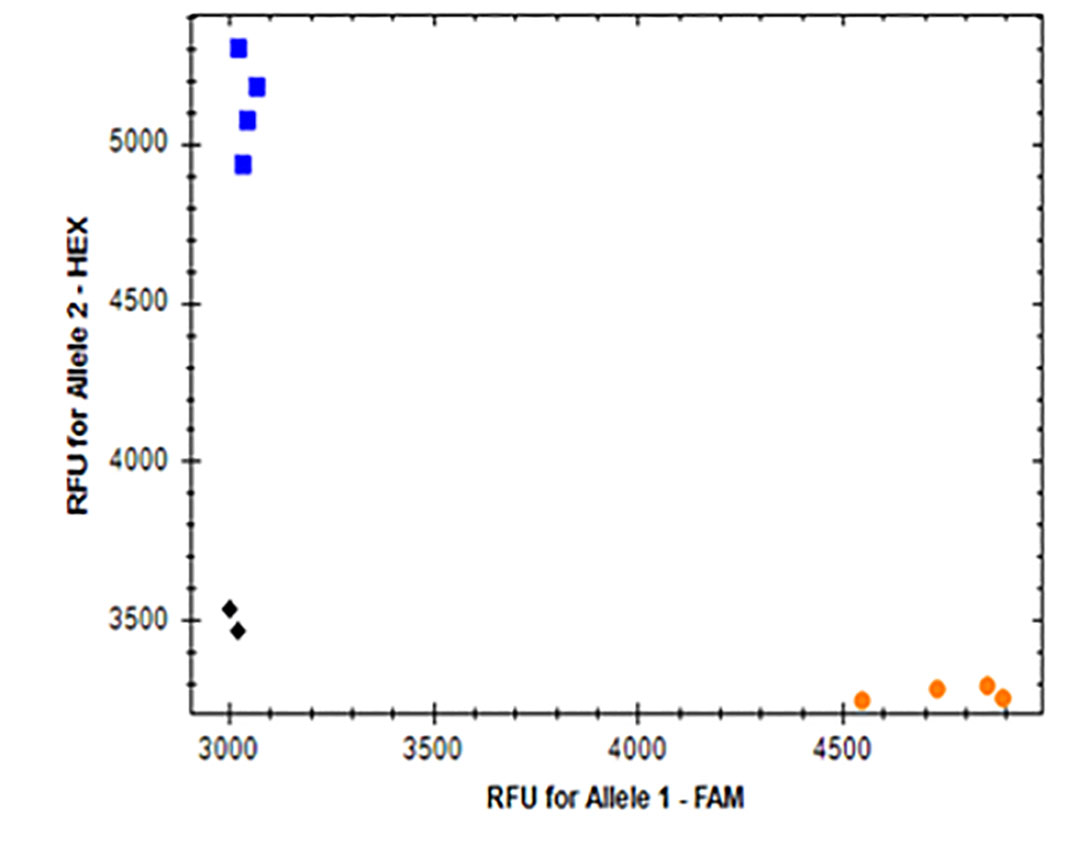
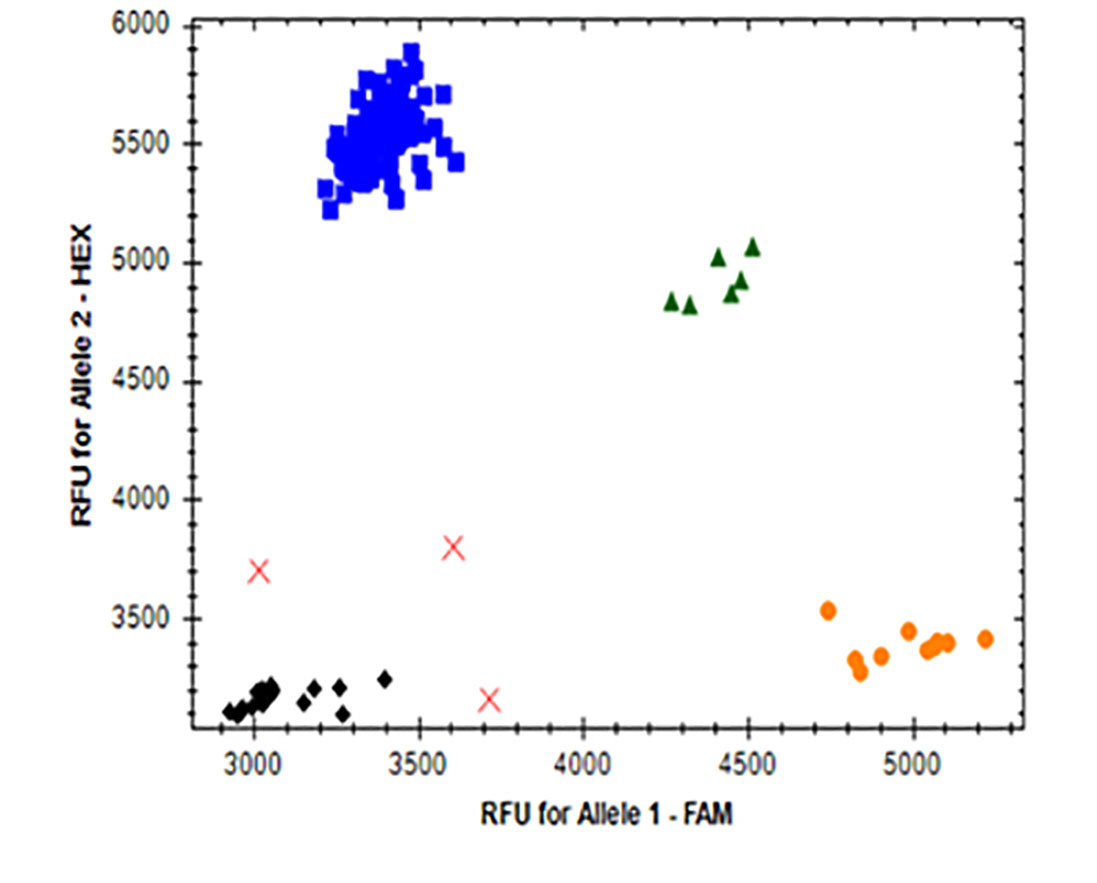
Figure 5. KASP marker for ACCase EMS mutation detection.
Allelic discrimination for ACCase-2A (A), ACCase-2B (B) and ACCase-2D (C) are shown as blue dots representing the ACCase mutant alleles, with the orange dots representing the ACCase wild-type alleles, and the green dots representing ACCase heterozygosity (one wild-type allele and one mutated allele). Black dots represent the non-template water control (NTC). Red X’s are undetermined samples. The X and Y axis show the fluorescent value of each FAM- and HEX-fused alleles, respectively.
