Gene Discovery, Transformation & Genomic Application
Newly cloned gene provides another key to winter wheat maturity
Flowering time or heading date is a critical trait that underlies wheat adaptation to diverse climates. Winter wheat varieties in Oklahoma are planted in the fall but do not initiate flowering until sufficiently cold-hardened and vernalized. While this provides a distinct advantage for freeze avoidance or for grazing winter wheat throughout the winter, this growth habit becomes increasingly vulnerable to global climate change as higher winter temperatures may result in incomplete vernalization. Therefore, the identification of genetic factors that subtly regulate flowering time in winter wheat is critical for wheat production and sustainable agriculture in an unpredictable or changing climate.
Duster and Billings were developed by the Oklahoma State University Wheat Improvement Team and were utilized in contrasting scenarios during their prime, usually a dual-purpose environment for Duster versus a grain-only environment for Billings. Duster’s ability to hold winter dormancy longer and flower later than Billings made it the preferred variety for dual-purpose wheat production. In previous studies, genetic variation was found in three known flowering time genes. The recessive vrn1, recessive vrn3 and PPD-sensitive alleles are often packaged together to regulate the developmental phases of winter wheat varieties for the southern plains. However, Duster and Billings have the same allele for each of these genes, indicating that there remains a missing regulatory genetic component(s) differentiating Duster and Billings. The immense challenge is to identify the gene(s) responsible for such a relatively minor difference of a few days in the field among these two winter wheat varieties.
A doubled haploid (DH) population was generated using Duster and Billings. Under controlled conditions, the developmental difference in flowering time was amplified from a few days in the field to 15 days (Fig. 1a). This facilitated mapping and identification of the missing genetic pieces. The DH population showed quantitative trait segregation in development (Fig. 1b). A major quantitative trait locus (QTL) was successfully mapped on chromosome 6A for flowering time in this population (Fig. 1c). This QTL was tightly and consistently associated with earliness of Billings or lateness of Duster in both controlled conditions and field conditions.
Through tremendous follow-up efforts over several years, WIT scientists have cloned a gene that regulates this difference in flowering in the Duster x Billings DH population. The target gene was named TaOGT1 for encoding a previously uncharacterized O-GlcNAc transferase (OGT) enzyme that is associated with accelerated flowering in Billings relative to Duster. Interestingly, TaOGT1 evokes a different regulatory mechanism than that associated with vernalization and photoperiod genes and thus expands the knowledge base for flowering time pathways in plant species.
To confirm that the cloned gene is the causal gene, WIT scientists used a gene gun to transform Duster with the specific allele from Billings. The resulting transgenic Duster showed the early flowering pattern expected of Billings. This natural variation in TaOGT1 relies on a 168-bp insertion in the regulatory region of the TaOGT1 gene in Billings (Fig. 1d). Diagnostic molecular markers were developed for the 168-bp insertion (Fig. 1e), which will be used to accelerate breeding novel wheat varieties adapted to Oklahoma.
TaOGT1 is a central regulator of the flowering pathway, including genes involved in vernalization, photoperiod and sugar metabolism (Fig. 2). The Oklahoma Wheat Research Foundation has enabled advancement of molecular technology from the triplelocus model to quadruple locus model for fine regulation of flowering time in Oklahoma wheat. This study was published in the journal Nature Communications. The research article was featured by Oklahoma State University in the press release OSU scientists make wheat gene discovery and has been widely cited by the USDA-National Institute of Food and Agriculture and by the International Service for the Acquisition of Agri-biotech Applications.
Discovery of a seedling death gene in wheat
A wheat plant, like all other living organisms, will die after completion of its life cycle. Two major types of cell death, apoptosis and necrosis, exist in higher organisms. Apoptosis is a term derived from ancient Greek that means "dropping off" or "falling off,” such as petals from flowers or leaves from trees. Necrosis, also derived from Greek, means "death, the stage of dying, the act of killing." Numerous genes causing death have been revealed in animals and humans, but no gene has been reported to underlie apoptosis or necrosis in wheat in previous studies.
Necrosis occurs in wheat as a type of death frequently observed in F1 hybrids from crosses between different wheat species. For example, when diploid T. tauschii (D genome, 14 chromosomes) is crossed with tetraploid T. turgidium L. (A and B genomes, 28 chromosomes) to generate synthetic hexaploid T. aestivum L. (A, B and D genomes, 42 chromosomes), hybrid necrosis may occur and is usually completely lethal in the absence of any pathogen or external stimulus. Hybrid necrosis has become a serious barrier that
- prohibits the combination of desirable traits from different genotypes even with the same number of chromosomes
- inhibits the development of mapping populations from specific parental combinations
- limits the utilization of heterosis in wheat
During a study on transgenic wheat, a mutant plant was discovered that died within two weeks after planting in the absence of any pathogen or external stimulus. The segregation ratio of 1:2:1 was observed for the number of wild type (completely viable seedlings), intermediate and mutant (necrotic) plants, fitting the single semi-dominant gene model (X21:2:1 = 1.308, p = 5.99, Fig. 3). The availability of surviving mutant plants in the heterozygous state facilitated cloning of the mutated gene. The point mutation was discovered in a gene that encodes a nucleotide-binding site leucine-rich repeat (NLR) protein. The mutation produces a protein devoid of a critical salt bridge in its structure (Fig. 3). Interestingly, the mutated NLR gene is orthologous to gene Sr35, which triggers the immune response to stem rust disease caused by Ug99, resulting in programmed cell death at the site of infection. This story has appeared in the prestigious journal Plant Physiology (Jia et al, 2020).
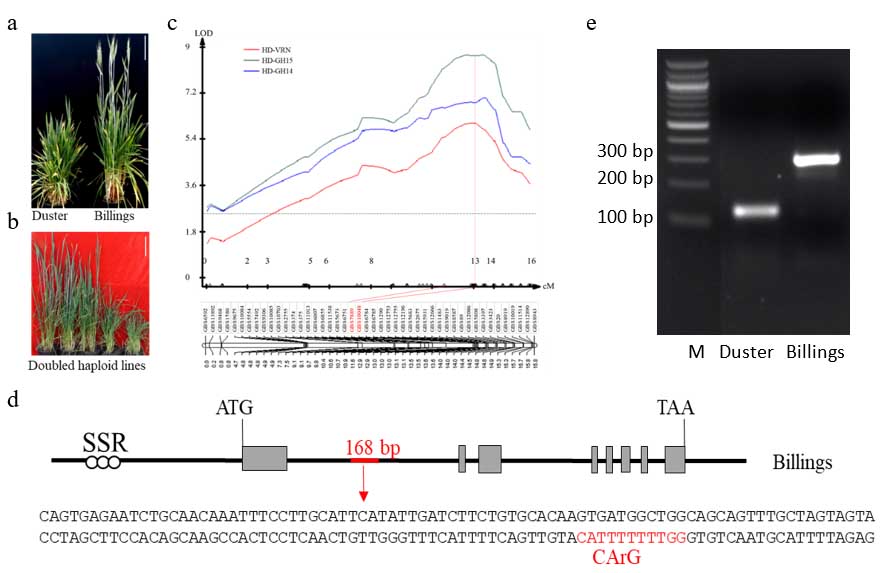
Figure 1. Mapping and cloning of TaOGT1 in wheat variety Billings.
- The two parental lines, Duster (left) and Billings, were grown in a greenhouse with controlled temperatures and long days without vernalization.
- Representative doubled haploid (DH) plants segregating for flowering time in the Duster × Billings DH population in the greenhouse and without vernalization. Scale bar = 15 cm in plates a and b.
- Mapping of QHd.osu-6A for flowering time. Phenotypic data were obtained from the DH population grown in the greenhouse and vernalized (HD-VRN) or not vernalized in 2014 (HD-GH14) and 2015 (HD-GH15). Two GBS markers, GBS7889 and GBS10048, linked with the QTL peak are indicated in red. The vertical dashed line indicates that the two GBS markers reside under the QTL peak. The x axis represents genetic distance (cM) within the genomic region covering QHd.osu-6A, and the y axis represents the log of the odds (LOD) value of QHd.osu-6A. The horizontal dashed line in the QTL curve of QHd.osu-6A represents a threshold LOD value of 2.5.
- Sequence of the 168-bp insertion in intron 1 of the Billings TaOGT1b allele. The CArG box is indicated in red, which is a conserved DNA target site of MADS-domain proteins, including VRN1 and TaVRT2.
- PCR marker for TaOGT1. Primers TaOGT1-F1 and TaOGT1-R1 amplified a 276 bp fragment from Billings but a 108 bp fragment from Duster due to a 168 bp indel variation (underlined) in the intron of TaOGT1.
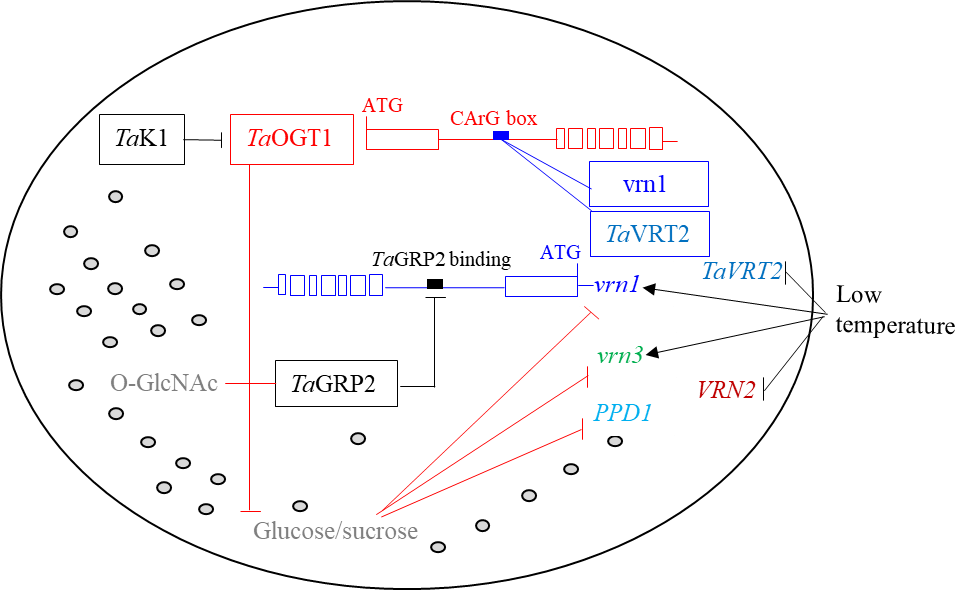
Figure 2. A model for the regulation of heading date by the TaOGT1-sugar pathway in winter wheat. Arrows indicate promotion and -| indicates repression. The genotypes of the winter wheat varieties Billings and Duster contain recessive vrn1 and vrn3 alleles and a photoperiod-insensitive allele, PPD1b. Italic names indicate genes, and non-italic names indicate proteins. Shaded gray circles represent the cytoplasmic matrix.

Figure 3. A gene causing seedling death in wheat. Segregating phenotypes of F2 plants are shown for the mutant plant x Bobwhite population (lower part). A salt bridge is diagrammed in the NLR protein (upper part).
