Unmanned Aerial Systems-Based High-Throughput Phenotyping 2023
Regular flights of the Stillwater location for the Dual-Purpose Observation Nursery by the OSU Wheat Improvement Team commenced after planting in September 2023 using a DJI Phantom 4 Multispectral Unmanned Aircraft System (UAS). Flight frequency was approximately once per week during the fall and twice per month during the winter. For each flight, multiple raw multispectral images were collected, covering the whole field with approximately 80% overlap between images (Figure 1).
The raw images were downloaded to a dedicated external drive and copied to a storage server at the OSU High-Performance Computing Center (HPCC) where image post-processing operations were performed (Figure 2). These raw images were post-processed using an algorithm called Structure from Motion (SfM). The SfM algorithm first geolocated each image using common features across images and Global Navigation Satellite System (GNSS) coordinates stored as exchangeable image file (EXIF) metadata within each image file (Figure 3).
The SfM algorithm also used the shared features to identify “tie points” (points within one image that match with points in another image), which were then used to generate a three-dimensional (3D) point cloud of the whole field trial (Figure 4). The points from the 3D point cloud were then connected to create a 3D mesh from which an orthomosaic multispectral image of the whole field was created. The orthomosaic image was then used to calculate a range of vegetation indices, including Normalized Difference Vegetation Index (NDVI; Figure 5).
Each sampling date generated approximately 500 GB of raw imagery and post-processed data.
Customized computer scripts for downloading data from drones, uploading and backing up imagery data to HPCC storage, and performing post-processing (orthorectification, mosaicking, and georeferencing) of imagery have been developed based on the R programming language and open-source software Open Drone Map. In addition, code has been written to extract plot-level summaries of relevant vegetation indices for extracting phenotypes to be used for subsequent selection. Figure 6 shows an overlay of polygons used for extracting plot-level phenotypes from orthomosaic imagery.

Figure 1. Multispectral imagery is captured using an Unmanned Aerial Systems (UAS) platform.

Figure 2. Example of raw multispectral image taken above of a portion of the field trial. Raw images are downloaded from the Unmanned Aerial System (UAS) to a computer for post-processing.
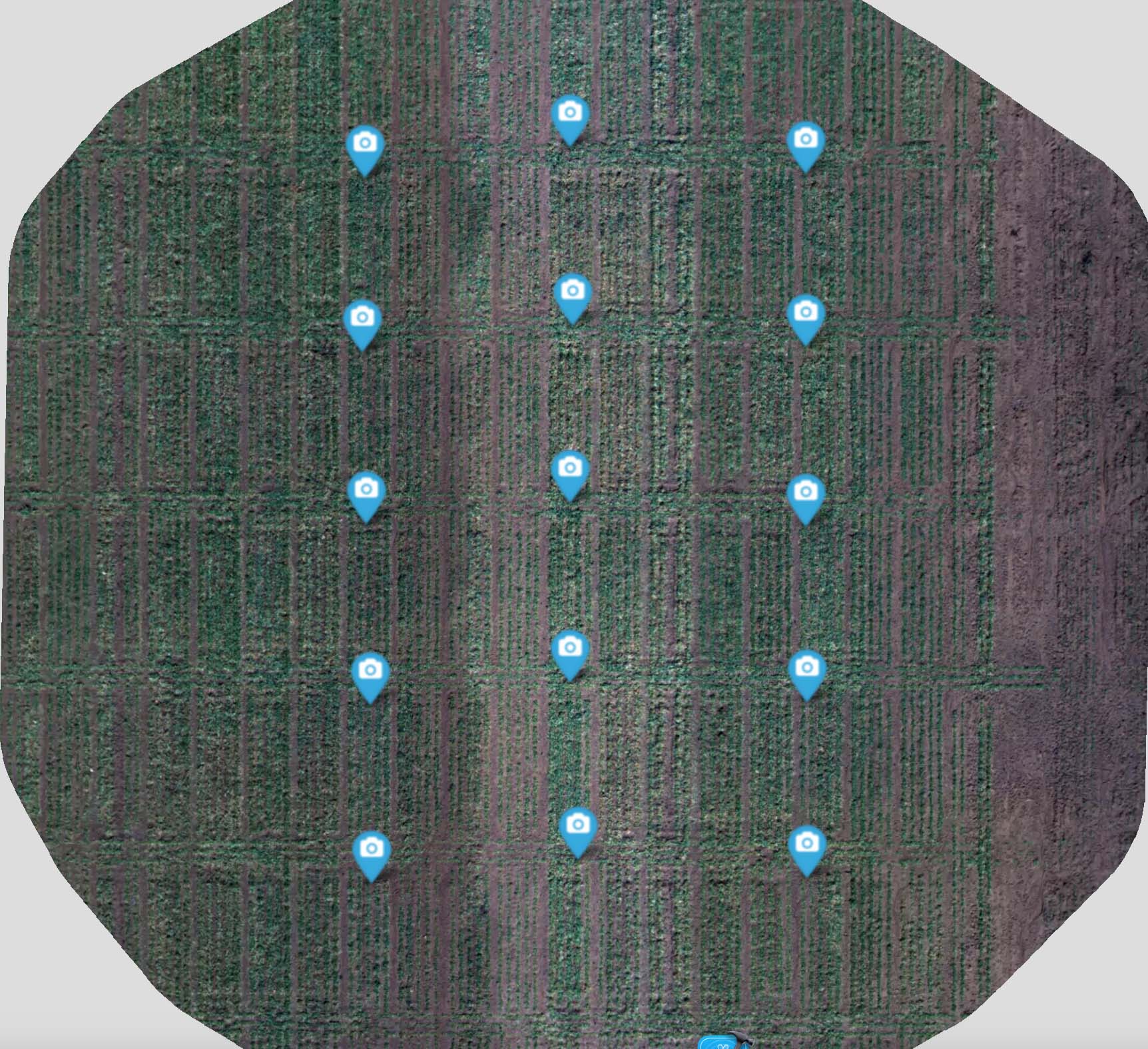
Figure 3. The points above indicate the geolocation of each raw image as determined using Global Navigation Satellite System (GNSS) coordinates stored in exchangeable image file (EXIF) metadata within each image and features extracted from each raw image by a Structure from Motion (SfM) algorithm.
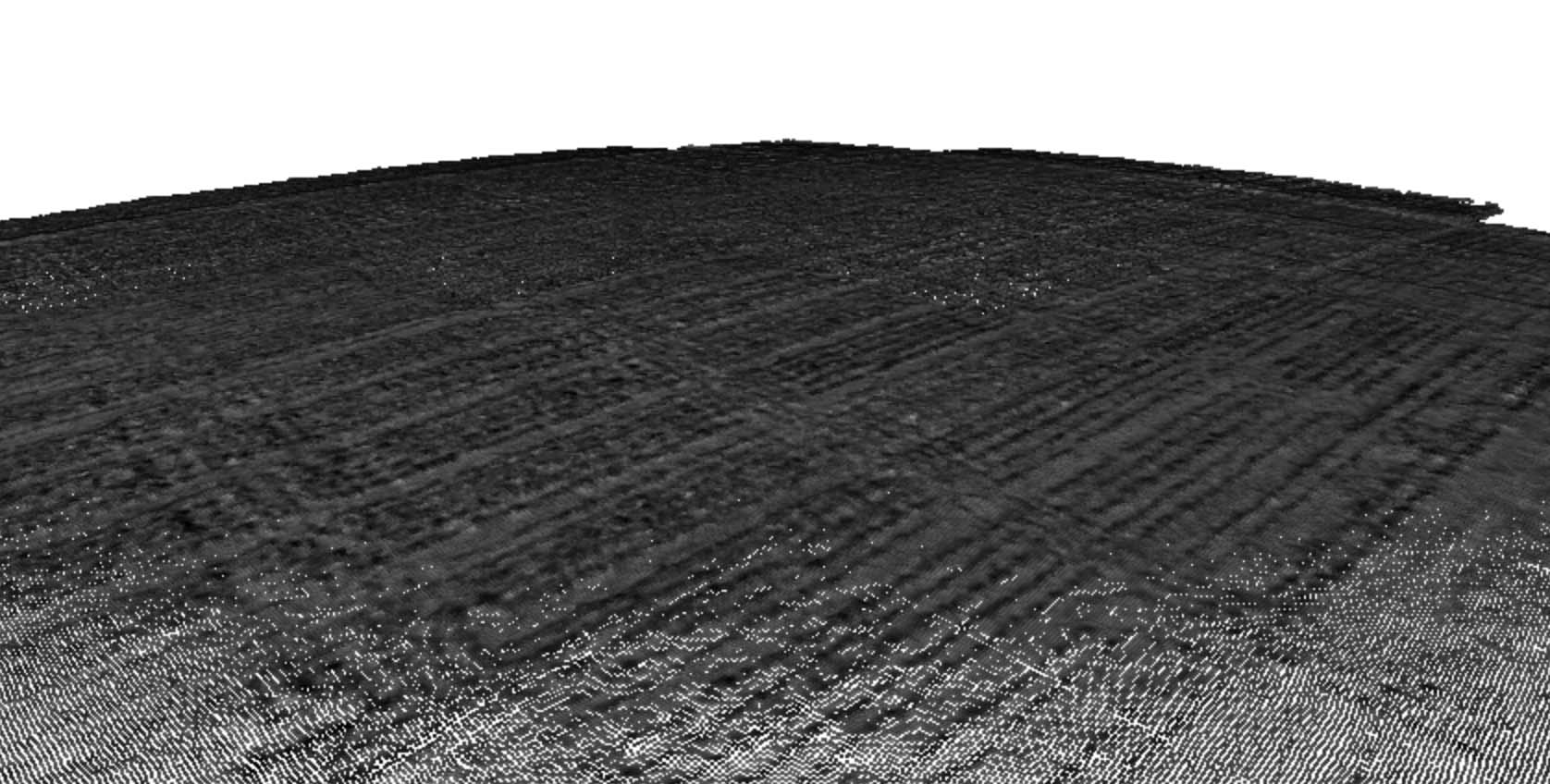
Figure 4. Three-dimensional point cloud of the field trial produced by a Structure from Motion (SfM) algorithm by combining features across multiple raw images.
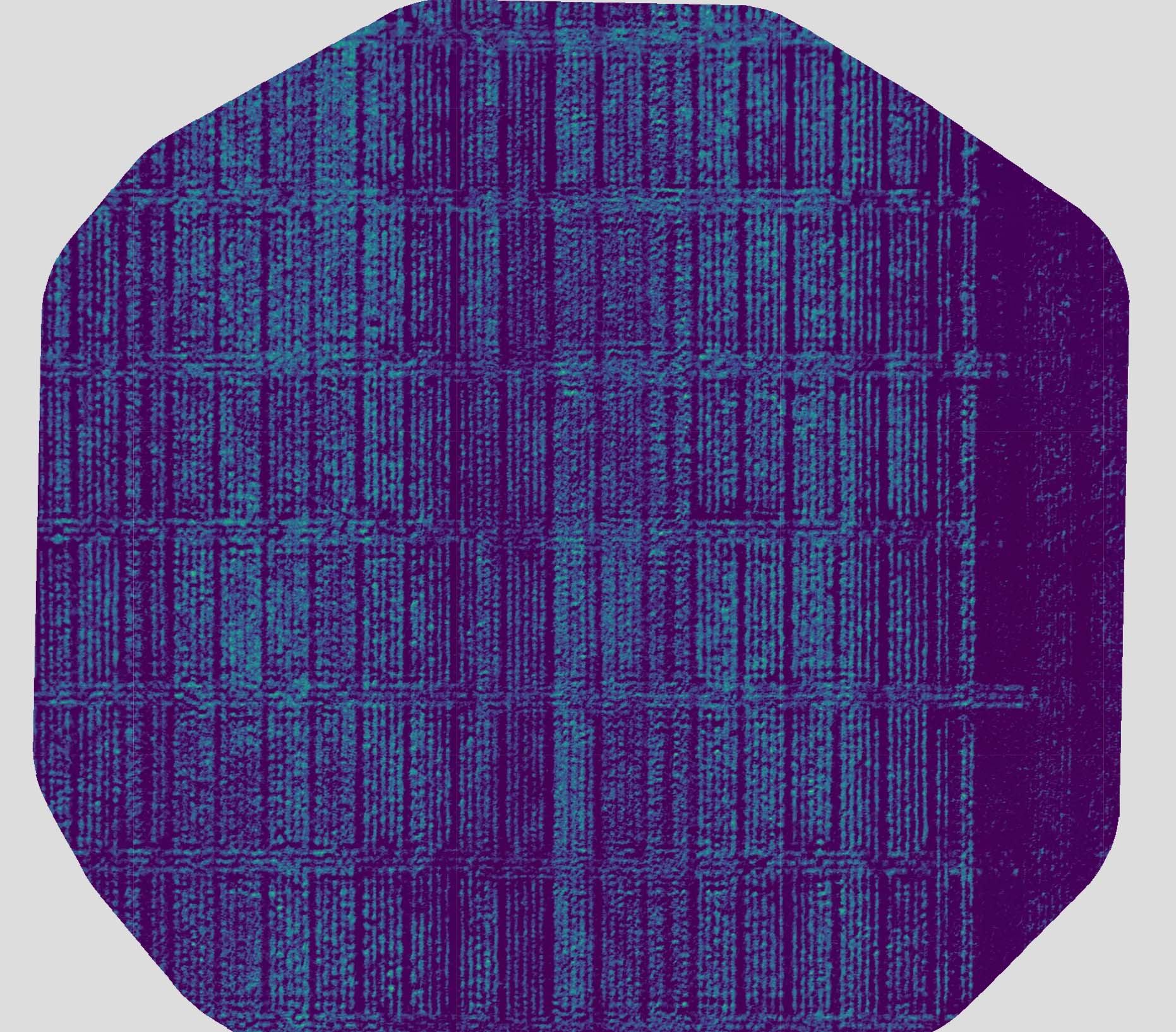
Figure 5. Normalized Difference Vegetation Index (NDVI) as calculated using the multispectral orthomosaic image generated from the mesh produced by connecting points from the three-dimensional point cloud produced by a Structure from Motion (SfM) algorithm.

Figure 6. Field plot polygons (yellow rectangles) that were used for extracting plot-level phenotypes overlaid onto an image of Normalized Difference Vegetation Index (NDVI) that was calculated from a multispectral orthomosaic image.
