Wheat Pathology Report for 2023
Disease evaluations of 2023 OSU wheat breeding lines
We determined the reactions of 983 Oklahoma State University breeding lines to fungal diseases, including leaf rust, powdery mildew, tan spot, and other leaf spotting diseases (spot blotch and Septoria nodorum blotch), and to viral diseases, including, barley yellow dwarf and wheat soil-borne mosaic/wheat spindle streak mosaic (WSBM/WSSM) (Table 1). Evaluated materials were at different stages in the breeding pipeline, including preliminary, intermediate, and elite breeding lines. As environmental conditions, such as heat and drought, may not favor natural disease development in the field, we conducted greenhouse evaluations for leaf rust, tan spot, and powdery mildew using artificial inoculations. In 2023, we generated a total of 22,548 disease data points for the OSU breeding lines.
| Disease | Test Condition | Number of breeding lines - Seedling Stage | Number of breeding ines - Adult-plant Stage |
|---|---|---|---|
| Leaf rust | Greenhouse | 456 | 456 |
| Powdery mildew | Greenhouse | 456 | 456 |
| Tan spot | Greenhouse | 136 | - |
| Leaf spotting diseases (spot blotch and Septoria nodorum blotch) | Field | - | 136 |
| Wheat soil-borne mosaic/wheat spindle stream mosaic | Field | - | 983 |
| Barley yellow dwarf | Field | - | 136 |
Symptoms of wheat diseases in Table 1, except Septoria nodorum blotch (SNB), can be found in the wheat pathology report for 2022. Septoria nodorum blotch, caused by the fungus, Parastagonospora nodorum, is manifested as a leaf blotch and glume blotch (Figures 1 & 2). SNB is becoming more prevalent in recent years in Oklahoma, occurring in four out of the last five years (2019 – 2023). Susceptibility to SNB is prevalent in the OSU breeding lines and varieties. OSU varieties Gallagher, Paradox, and Breadbox are highly susceptible to SNB, while Big Country and Uncharted are resistant.

Figure 1. Septoria nodorum blotch on wheat leaves (Morris, Okmulgee County, May 16, 2023).

Figure 2. Septoria nodorum blotch on glumes and awns of the wheat cultivar Breadbox (Stillwater, Payne County, May 17, 2023).
In addition to OSU breeding lines, 566 U.S. Great Plains hard winter wheat breeding lines from 2023 USDA-Agricultural Research Service regional nurseries were evaluated to WSBM/WSSM in Stillwater. WSBM/WSSM symptoms in the Stillwater nursery were reduced on the susceptible check Vona and were highly variable across the nursery in recent years. Hence, a molecular marker that detects the presence or absence of WSBM resistance gene Sbm1 in OSU elite lines was used in marker-assisted selection. Disease testing in the greenhouse and field will continue to enable line advancement in the OSU wheat breeding program. OSU released varieties have progressed toward good disease resistance and reduced dependence on fungicides. Recently released OSU varieties with best disease protection include Big Country, Uncharted, High Cotton, Skydance, Butler’s Gold, Smith’s Gold, OK Corral, Doublestop CL+, and Strad CL+. Breakthrough, targeted for the Oklahoma panhandle, does not require the same level of fungal disease protection but instead features highly effective levels of viral resistance to the wheat streak mosaic/Triticum mosaic complex. Successors to Breakthrough will be addressed in this report for the 2024 crop season.
Understanding the underlying genetics of rust resistance in OSU hard winter wheat
Leaf rust and stripe rust are the most devastating wheat diseases globally. Over the last five years (2018-2022), average annual yield loss in Oklahoma due to leaf rust and stripe rust were estimated at $20 million and $15 million, respectively. Therefore, we focused on identifying leaf rust and stripe rust resistance sources, mapping rust resistance genes, and developing diagnostic markers for use in marker-assisted selection.
Hard winter wheat association mapping panel:
We created a germplasm collection of 626 breeding lines and cultivars from the OSU wheat breeding program. This collection includes
- 38 elite breeding lines from 2022
- 15 recent OSU varieties
- 533 doubled-haploid lines selected from 14 bi-parental crosses
- a historical OSU panel of 40 lines and varieties
This collection was evaluated at the seedling stage in the greenhouse with U.S. leaf rust and stripe rust pathogen races and evaluated at the adult plant stage to leaf rust in Stillwater and to stripe rust in Chickasha (Figure 3). We observed higher frequencies of lines with rust resistance at the adult plant stage compared with the seedling stage. This suggests the presence of adult plant resistance (APR) in OSU breeding lines. In the fall of 2023, this germplasm was planted in rust nurseries in Oklahoma (Stillwater and Chickasha), Kansas (Rossville and Hutchinson), and Washington (Pullman) for rust evaluations in Spring 2024.
This germplasm was genotyped at the USDA-ARS Central Small Grains Genotyping Lab in Manhattan, Kansas. A total of 30,524 high-quality single nucleotide polymorphism (SNP) markers, distributed across the whole wheat genome, were retained for future analysis to identify genomic regions associated with leaf rust and stripe rust resistance.
Figure 3a-d. Rust responses of 626 OSU hard winter wheat lines and varieties. Susceptible lines have higher values of infection types, disease severity, and coefficient of infection. Leaf rust coefficient of infection of the susceptible check OK Bullet was equal to 34 on average (moderate disease pressure). Stripe rust severity of the susceptible check Pete was equal to 80% on average (high disease pressure).
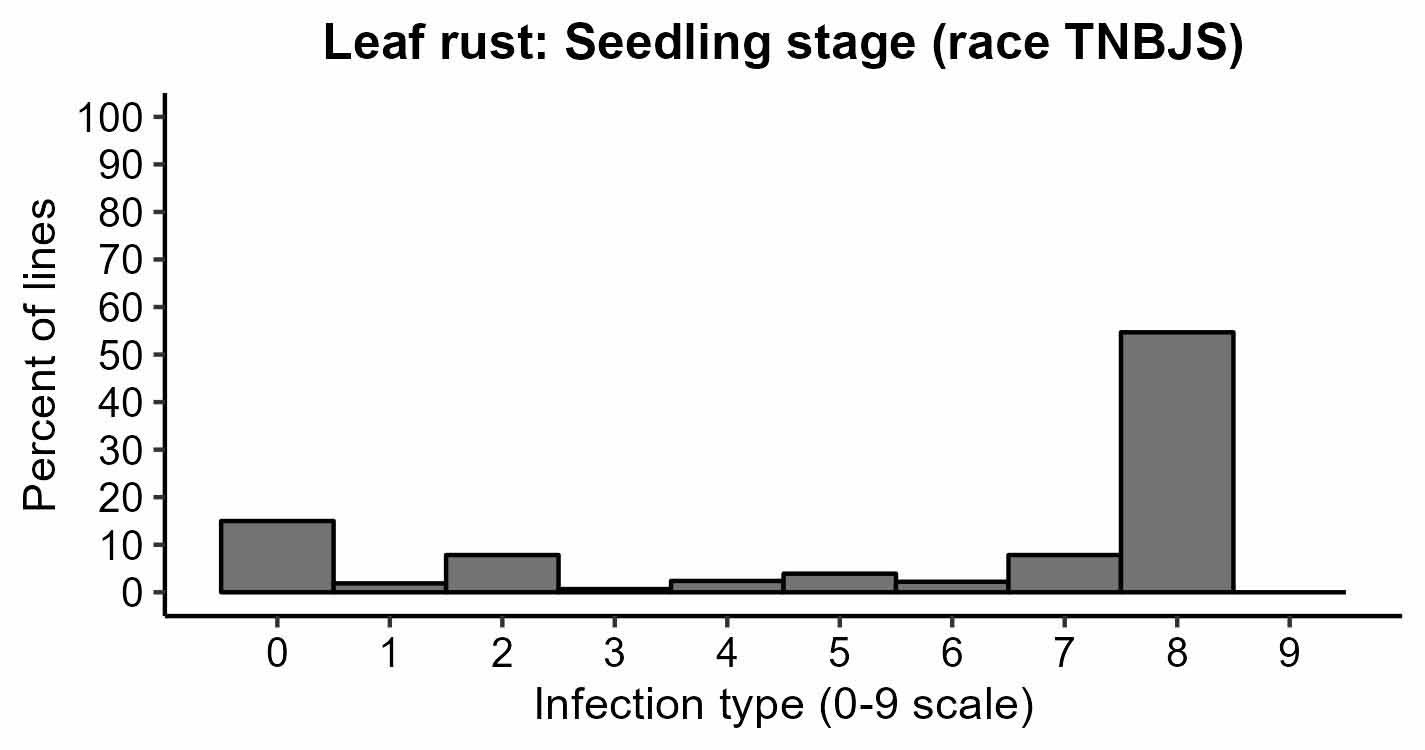
Figure 3a. Leaf rust: Seedling stage (race TNBJS).
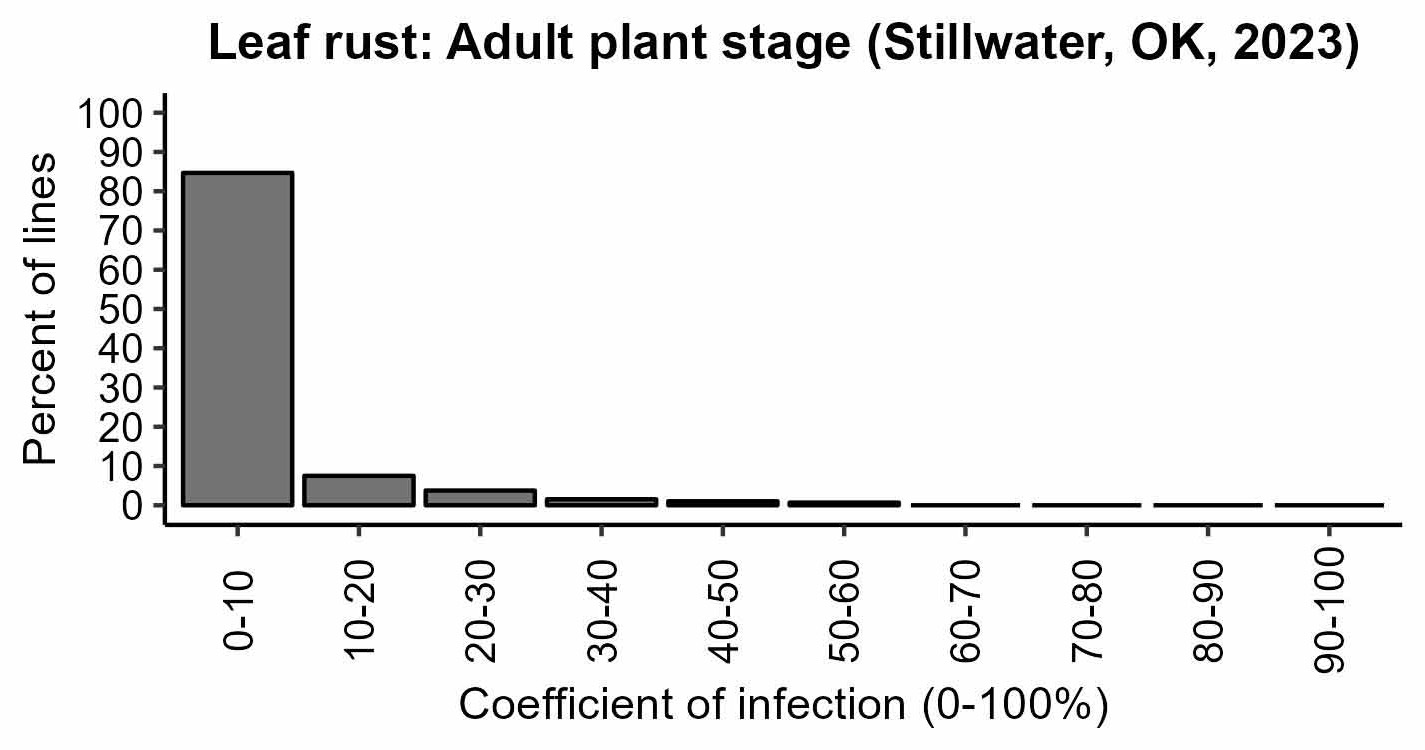
Figure 3b. Leaf rust: Adult plant stage (Stillwater, OK, 2023).
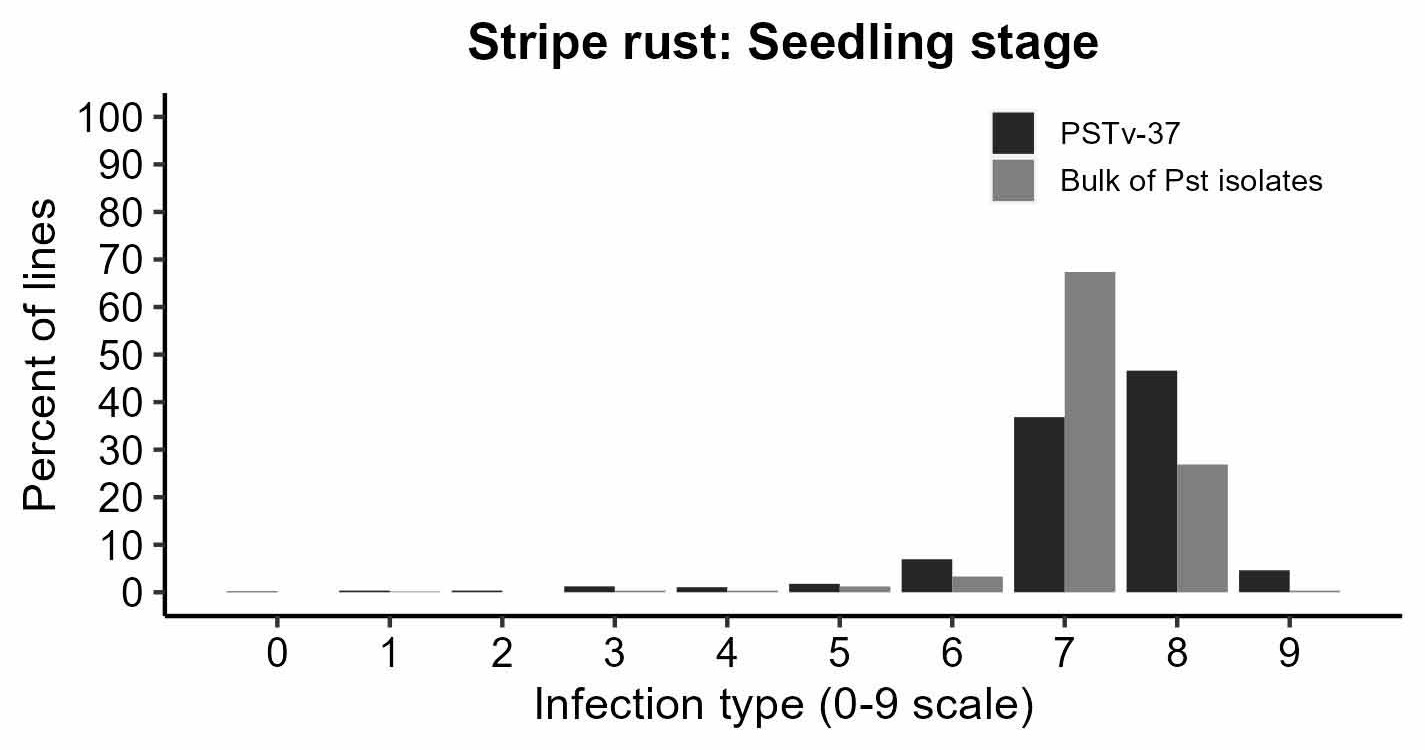
Figure 3c. Stripe Rust: Seedling stage
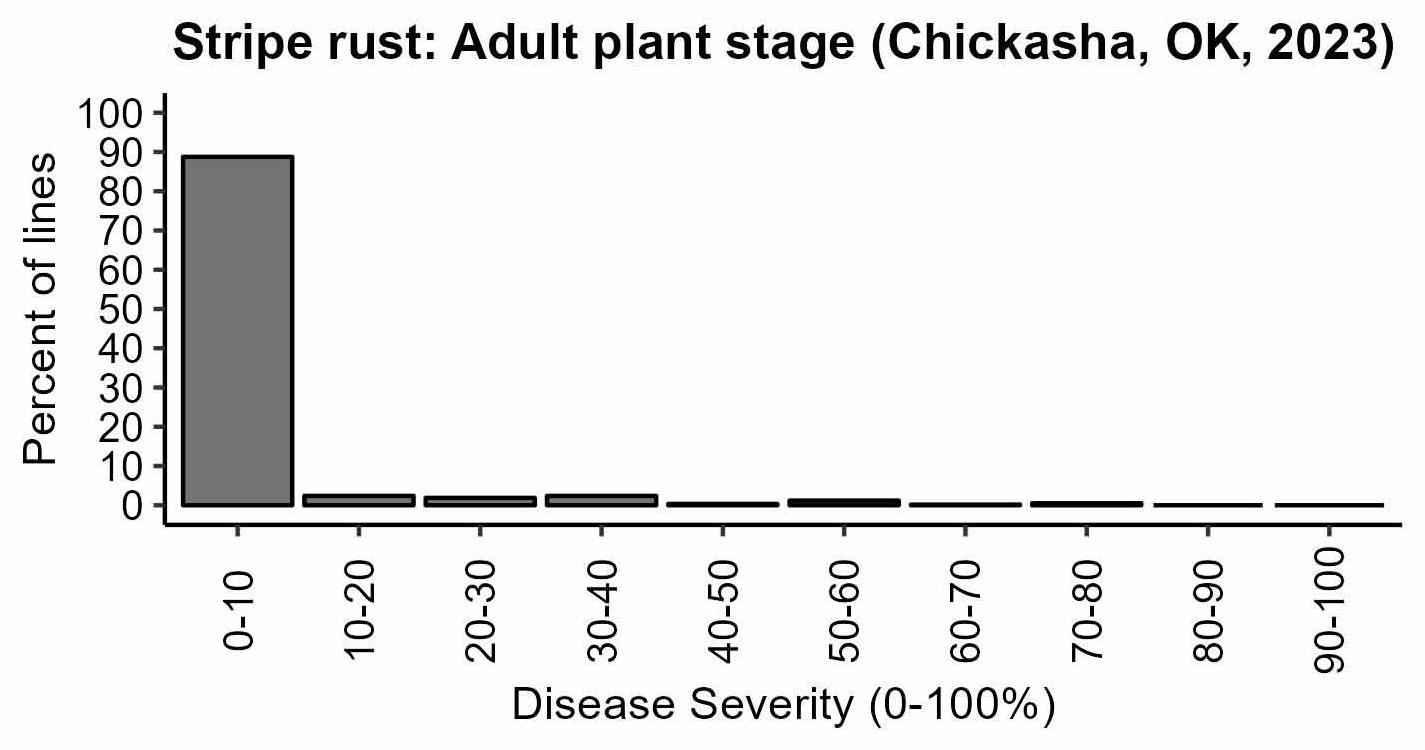
Figure 3d. Leaf rust: Adult plant stage (Chickasha, OK, 2023).
Hard winter wheat bi-parental mapping populations:
To map and identify adult plant resistance (APR) genes to stripe rust in OSU varieties Gallagher, Baker’s Ann, and Green Hammer, three bi-parental populations, were planted in fall 2023 in Oklahoma (Chickasha), Kansas (Rossville), and Washington (Pullman and Mt. Vernon) for stripe rust evaluation in Spring 2024. We are also developing a bi-parental population Big Country/Jagalene that should be available for phenotyping and genotyping in Spring 2024 to characterize leaf rust and stripe rust resistance genes in the OSU variety Big Country. The bi-parental populations used in this project include:
Gallagher/Iba:
170 F4:6 recombinant inbred lines (RILs) were developed from a cross between the two Oklahoma hard red winter wheat varieties Gallagher and Iba. Gallagher and Iba are half-sibs released in 2012. However, Gallagher offers stronger protection at the adult plant stage against stripe rust compared to Iba. Gallagher is moderately resistant, whereas Iba has an intermediate reaction to stripe rust. Gallagher was found to carry the resistance genes TaXA21-A1 (also present in its parent ‘Duster’) and Lr46/Yr29. Iba was found to carry the resistance genes Lr34/Yr18, TaXA21-A1, and Lr46/Yr29. Therefore, we hypothesize that other unknown APR gene(s) against stripe rust should be present in Gallagher to explain its higher protection compared to Iba.
OK12D22004-016/Baker’s Ann:
A doubled-haploid (DH) population of 125 DH lines was developed from a cross between the OSU breeding line OK12D22004-016 and the OSU variety Baker’s Ann. Baker’s Ann was released in 2018 and is highly resistant to stripe rust at the adult plant stage. Based on diagnostic molecular markers for the characterized stripe rust APR genes (i.e., Lr34/Yr18, Lr46/Yr29, Yr36, and Lr67/Yr46), Baker’s Ann was found to carry Lr46/Yr29. However, the high level of protection in Baker’s Ann cannot be explained by a single APR gene that provides partial resistance. Thus, we hypothesize that Baker’s Ann should carry other unknown stripe rust APR genes.
Green Hammer/Lonerider:
109 DH lines were developed from a cross between the two Oklahoma varieties Green Hammer and Lonerider. Green Hammer was released in 2018 and is moderately resistant to stripe rust at the adult plant stage, whereas Lonerider is moderately susceptible. Based on molecular markers for the characterized stripe rust APR genes (i.e., Lr34/Yr18, Lr46/Yr29, Yr36, and Lr67/Yr46), Green Hammer does not carry any of these known APR genes. Thus, the genetic factor(s) underlying Green Hammer’s stripe rust APR is unknown.
Big Country/Jagalene:
A total of 160 DH lines were available in fall 2023. Seeds of the DH lines are being increased to enable rust evaluations and genotyping. Big Country is resistant to multiple diseases, including leaf rust and stripe rust. Based on moderately diagnostic molecular markers for known leaf rust and stripe rust resistance genes (i.e., Lr18, Lr19, Lr21, Lr24, Lr34/Yr18, Lr37/Yr17, Lr42, Lr46/Yr29, Lr67/Yr46, Lr68, Lr77, Lr78, Yr5, Yr15, Yr36, and QYr.tamu-2B), Big Country was found to carry the leaf rust APR gene Lr68 (partial resistance) and the ASR leaf rust/stripe rust resistance gene Lr37/Yr17 (not effective against current U.S. races). Therefore, Big Country should carry other unknown leaf rust resistance gene(s). Furthermore, Big Country stripe rust resistance was primarily due to APR genes that are yet to be discovered. In contrast to Big Country, Jagalene is susceptible to both leaf rust and stripe rust and carries Lr24 and Lr37/Y17, which are not effective against current U.S. races.
Association analysis to identify genomic regions associated with leaf rust and stripe rust resistance in U.S. Great Plains hard winter wheat
Currently, 22% and 26% of hard winter wheat (HWW) varieties grown in the Great Plains were rated as highly resistant or moderately resistant to stripe rust and leaf rust, respectively. Therefore, more varieties with durable rust resistance are needed for the Great Plains. In addition, the genetic basis of leaf rust and stripe rust resistance in U.S. HWW remains largely unknown. The identification of rust resistance genes will enable breeders to incorporate and pyramid multiple and diverse sources of rust resistance genes using marker-assisted selection, which will promote gene stewardship in future HWW varieties.
In 2022-2023, we initiated a multi-state project in collaboration with the USDA-ARS to identify rust resistance genes in current U.S. Great Plains HWW varieties originated from multiple breeding programs across 13 states. We selected 459 breeding lines and varieties from the 2021 and 2022 Northern Regional Performance Nursery (NRPN), Southern Regional Performance Nursery (SRPN), and the Regional Germplasm Observation Nursery (RGON). In 2023, this germplasm was evaluated for reaction to leaf rust and stripe rust at the adult plant stage in the field in Oklahoma (Stillwater and Chickasha), Kansas (Rossville), and Washington (Pullman and Mt. Vernon). We also conducted seedling tests in the greenhouse using five U.S. leaf rust pathogen races (i.e., MNPSD, MBDSD, TNBJS, MPPSD, and MJBJG) and five U.S. stripe rust pathogen races (PSTv-4, PSTv-14, PSTv-37, PSTv-40, and PSTv-52). Sources of seedling resistance (also known as all-stage resistance (ASR) or race-specific resistance) and APR (also known as non-race specific resistance) were identified in this germplasm to both leaf rust and stripe rust.
At the seedling stage, 39- 59% of the lines were moderately to highly resistant (depending on the race), whereas 66% of the lines were highly resistant to leaf rust (coefficient of infection ≤ 20%) at the adult plant stage in Stillwater. However, leaf rust pressure was not high in 2023 in Oklahoma due to drought. Therefore, additional leaf rust testing will be performed in a Stillwater leaf rust nursery in Spring 2024. At the seedling stage, low frequencies of moderately to highly resistant lines to stripe rust were identified (4-7% depending on the race). Stripe rust resistance in Great Plains HWW was primarily due to APR (32-78% of lines were moderately to highly resistant, depending on the location). We identified a total of 11 breeding lines from the 2022 RGON that were highly resistant across all tested races at the seedling stage (infection type ≤ 3) and across different locations at the adult plant stage (infection type ≤ 3 and disease severity ≤ 20%) (Table 2). These lines with broad-spectrum resistance to stripe rust can be used to breed for ASR, which is rare in current Great Plains HWW. We also identified 140 lines that are sources of APR.
| Line name | Origin | Known stripe rust resistance genes based on diagnostic markers |
|---|---|---|
| TX18DH313 | Texas | Yr5, QYr.tamu-2B |
| KS21U7494.H5.C1 | Kansas | Yr5, Yr15, QYr.tamu-2B |
| TX18DH305 | Texas | Lr37/Yr17, Yr5, Yr15, QYr.tamu-2B |
| TX18DH266 | Texas | QYr.tamu-2B, Lr46/Yr29 |
| KS21HD144 | Kansas | Lr37/Yr17, Yr15, Lr46/Yr29 |
| KS21HD147 | Kansas | Lr37/Yr17, Yr5, Lr46/Yr29 |
| KS21U7494.G14.C6 | Kansas | Yr5, Yr15, QYr.tamu-2B, Lr46/Yr29 |
| KS21U7494.H1.B8 | Kansas | Yr5, Yr15, QYr.tamu-2B, Lr46/Yr29 |
| TX18DH303 | Texas | Lr37/Yr17, Yr5, QYr.tamu-2B, Lr46/Yr29 |
| KS21U7266.E1.B2 | Kansas | Lr37/Yr17, Yr15, QYr.tamu-2B, Lr34/Yr18, Lr46/Yr29 |
| CO19D304R | Colorado | No known genes |
This germplasm was genotyped by USDA-ARS using moderately diagnostic molecular markers for known leaf rust and stripe rust resistance genes (i.e., Lr18, Lr19, Lr21, Lr24, Lr34/Yr18, Lr37/Yr17, Lr42, Lr46/Yr29, Lr67/Yr46, Lr68, Lr77, Lr78, Yr5, Yr15, Yr36, and QYr.tamu-2B). The studied germplasm was found to carry Lr21, Lr24, Lr34/Yr18, Lr37/Yr17, Lr46/Yr29, Lr68, Lr77, Lr78, Yr5, Yr15, and QYr.tamu-2B. This germplasm was also genotyped at USDA-ARS using a high-density single nucleotide polymorphism (SNP) marker platform, and 9,858 SNP markers distributed across the wheat genome were generated. Genome-wide association studies (GWAS) were performed using phenotypic rust data and genotypic data. A total of 26 SNP markers were significantly associated with leaf rust responses at the seedling stage (Figure 4). The SNP marker S3D_150055006 on chromosome 3D at 150 Mb (million base pair) was associated with leaf rust response to all five races. The SNP marker S6A 611716646 on chromosome 6A at 612 Mb was associated with leaf rust response to three races (i.e., MBDSD, MJBJG, and TNBJS) (Figure 4). These two markers are currently prioritized for the development of user-friendly markers – Kompetitive Allele-Specific PCR (KASP) markers – that can be used in high-throughput marker-assisted selection. A total of 12 SNP markers were associated with leaf rust response at the adult plant stage in Stillwater. The SNP marker S3A_568983726 on chromosome 3A at 569 Mb was the most significant marker associated with leaf rust response at the adult plant stage (Figure 5).
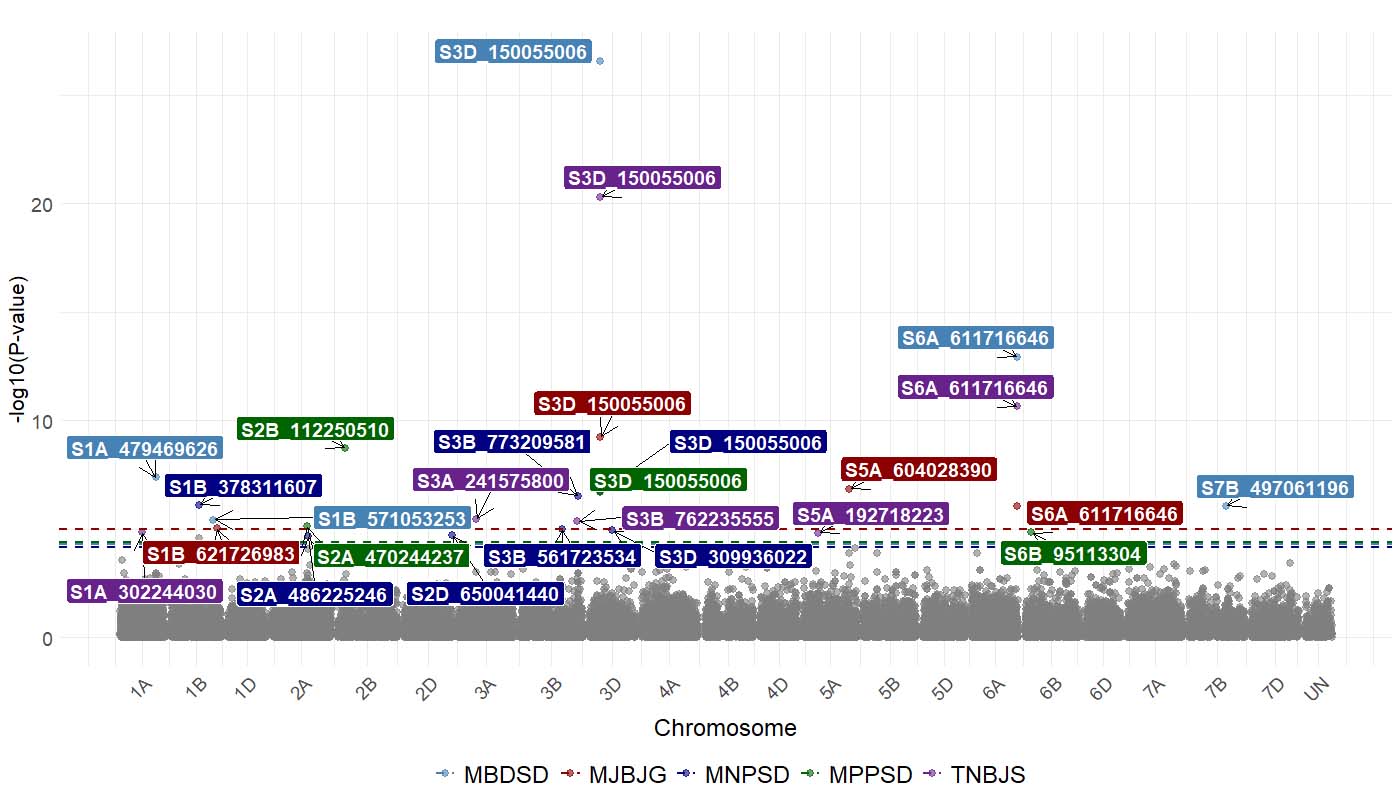
Figure 4. Molecular markers associated with leaf rust seedling response in Great Plains hard winter wheat. Different colors correspond to associated markers with leaf rust response to different races. The markers are designated with the chromosome name and a number that corresponds to the physical position in base pairs on the Chinese Spring reference genome IWGSC v2.1.
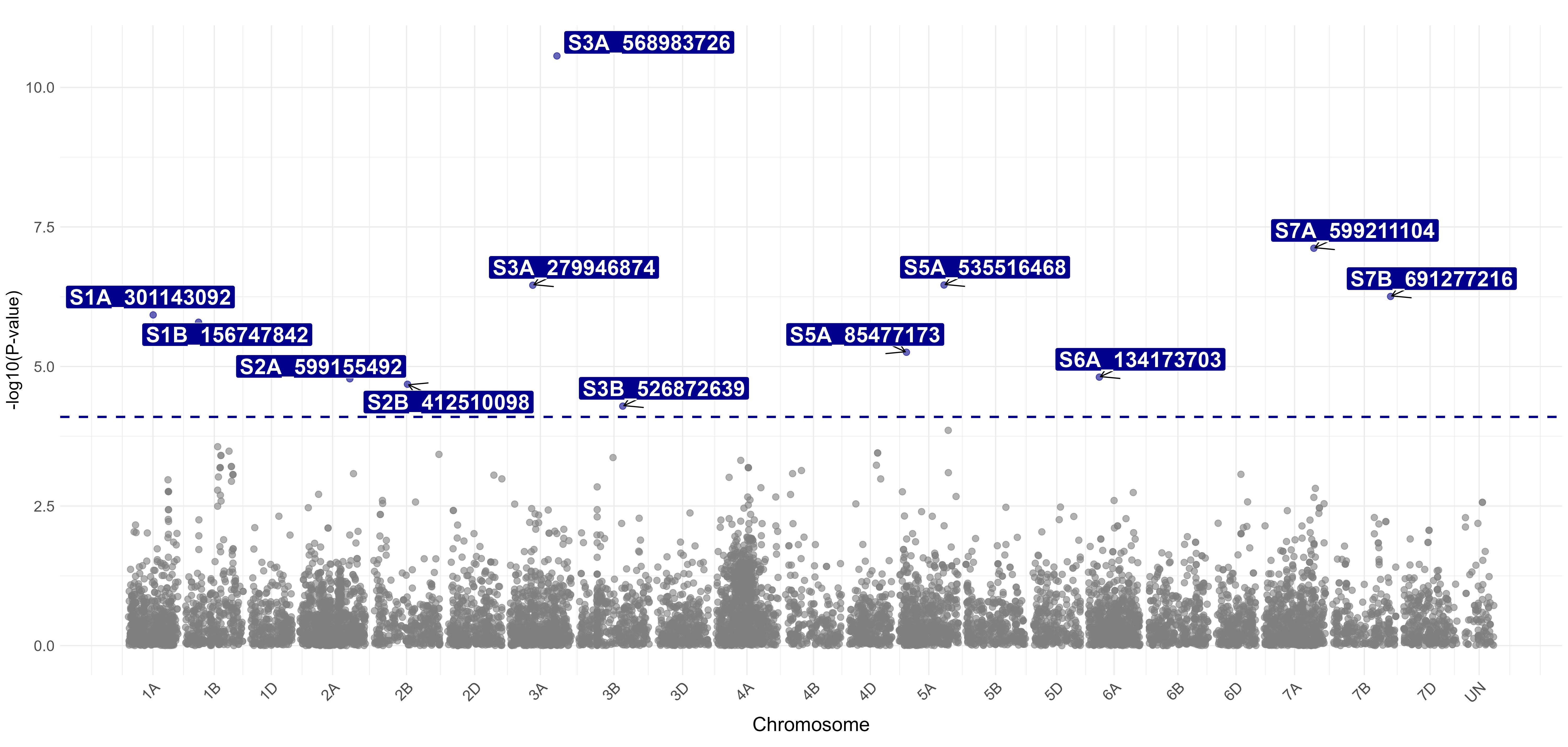
Figure 5. Molecular markers associated with leaf rust response at the adult-plant stage in Stillwater. The markers are designated with the chromosome name and a number that corresponds to the physical position in base pairs on the Chinese Spring reference genome IWGSC v2.1.
For stripe rust, GWAS identified 46 significant SNP markers associated with stripe rust response at the seedling stage (Figure 6). Four of these markers (S1B_141655194, S2B_458606839, S2B_598744752, and S2B_706506028) were associated with seedling response to at least three of the five tested stripe rust pathogen races. A total of 16 significant markers were associated with infection types and disease severity at the adult plant stage across field environments (Figure 7). Six of these markers, S2A_480477702, S2A_638603325, S2B_48062121, S3A_261175449, S4B_571886653, and S7D_518590833 had relatively larger effects across environments and will be prioritized for the development of KASP markers. A few of the associated SNP markers with stripe rust response were in proximity to known Yr genes, including Yr5, Yr10, Yr26, Yr53, Yr62, Yr68, Yr84, and Yr85. Most of the genomic regions that are associated with leaf rust and stripe rust responses in this study were not previously known. This provides an opportunity to enhance the deployment of diverse and durable sources of rust resistance in HWW and thus fulfill one of WIT’s top objectives for wheat improvement.
Figure 6a-e. Molecular markers associated with the stripe rust seedling response in Great Plains hard winter wheat. Associated markers that are in proximity to characterized stripe rust resistance (Yr) genes in wheat are indicated with the arrows.
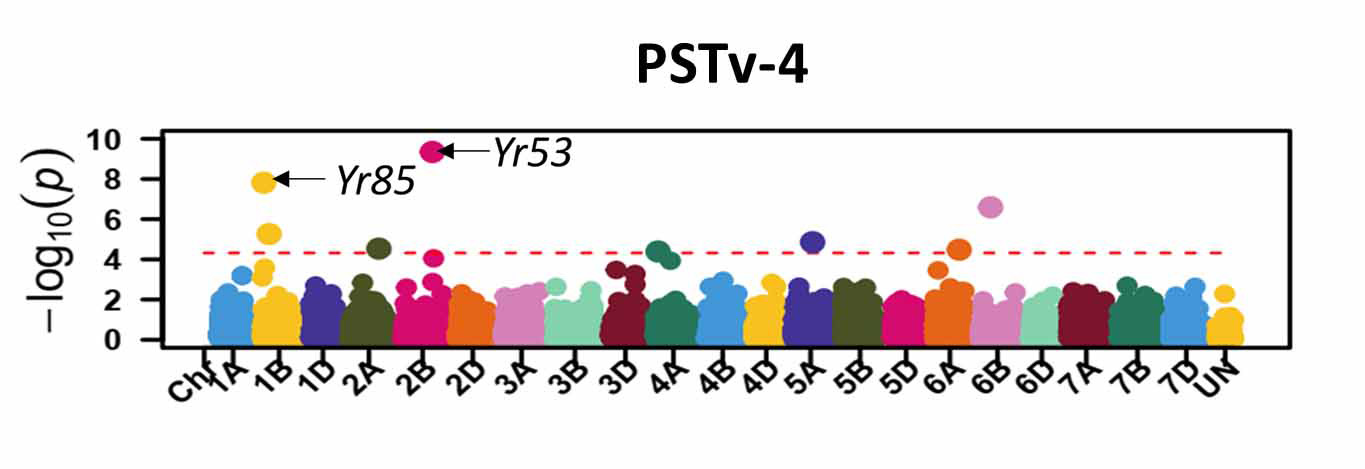
Figure 6a. PSTv-4.
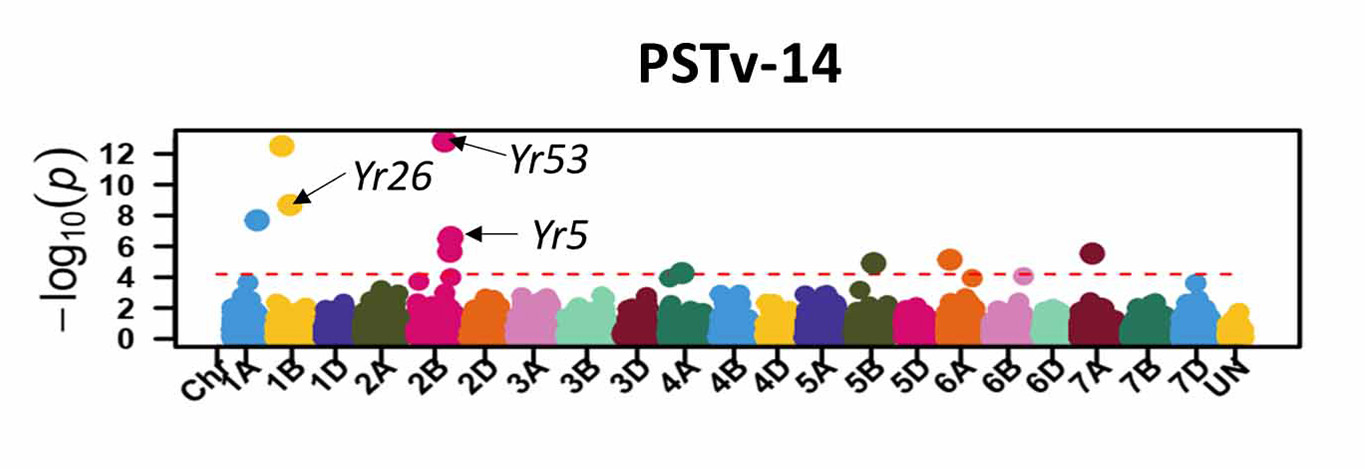
Figure 6b. PSTv-14.
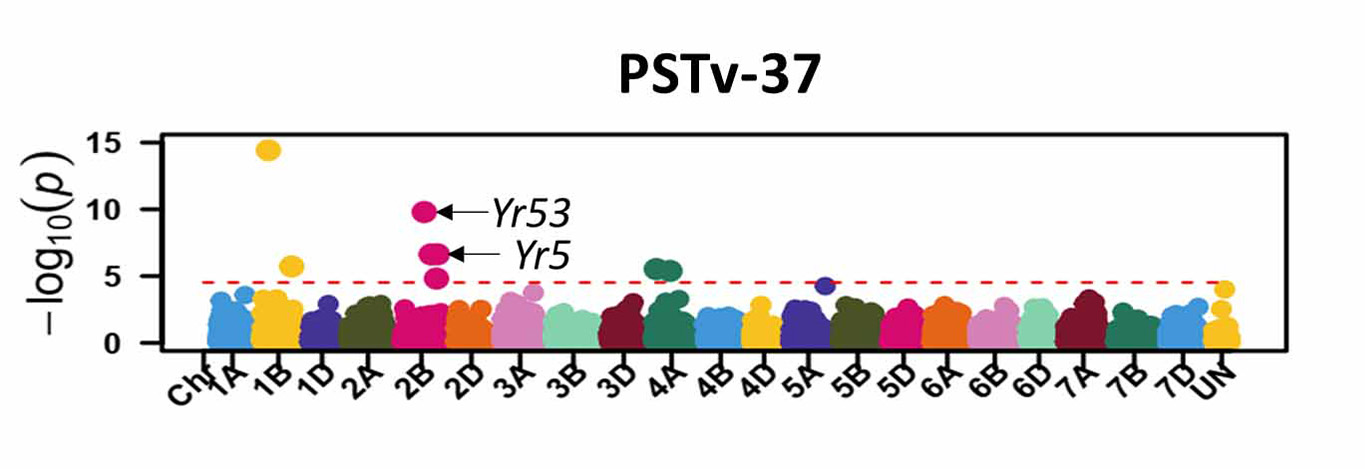
Figure 6c. PSTv-37.
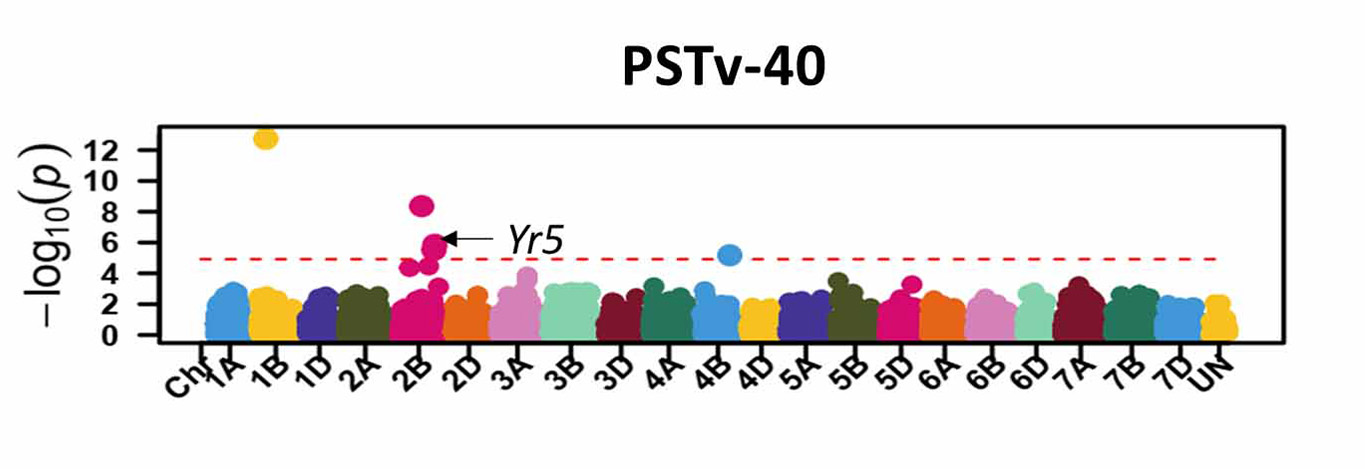
Figure 6d. PSTv-40.
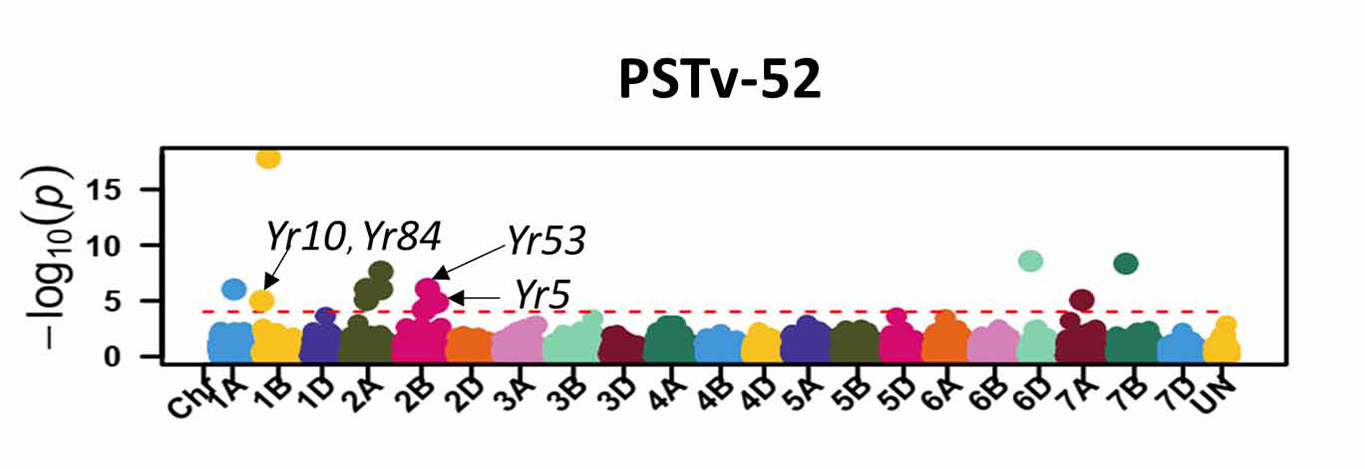
Figure 6e. PSTv-52.
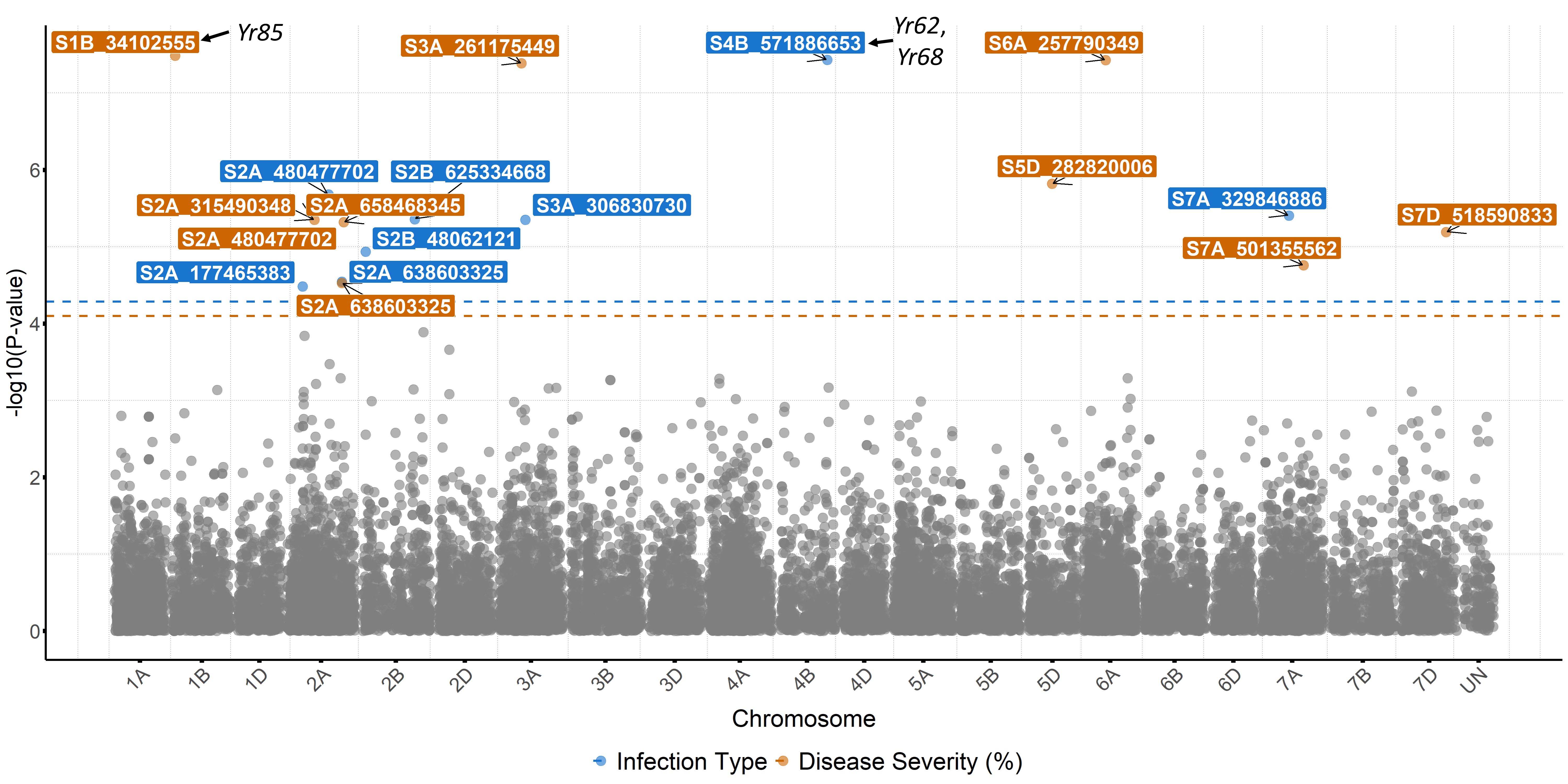
Figure 7. Molecular markers associated with stripe rust response at the adult plant stage across four field locations in Oklahoma, Kansas, and Washington. Different colors correspond to associated markers with the best linear unbiased predictions for infection type (blue) and disease severity (red) across locations. The markers are designated with the chromosome name and a number that corresponds to the physical position in base pairs on the Chinese Spring reference genome IWGSC v2.1. Associated markers that are in proximity to characterized stripe rust resistance (Yr) genes in wheat are indicated with arrows.
